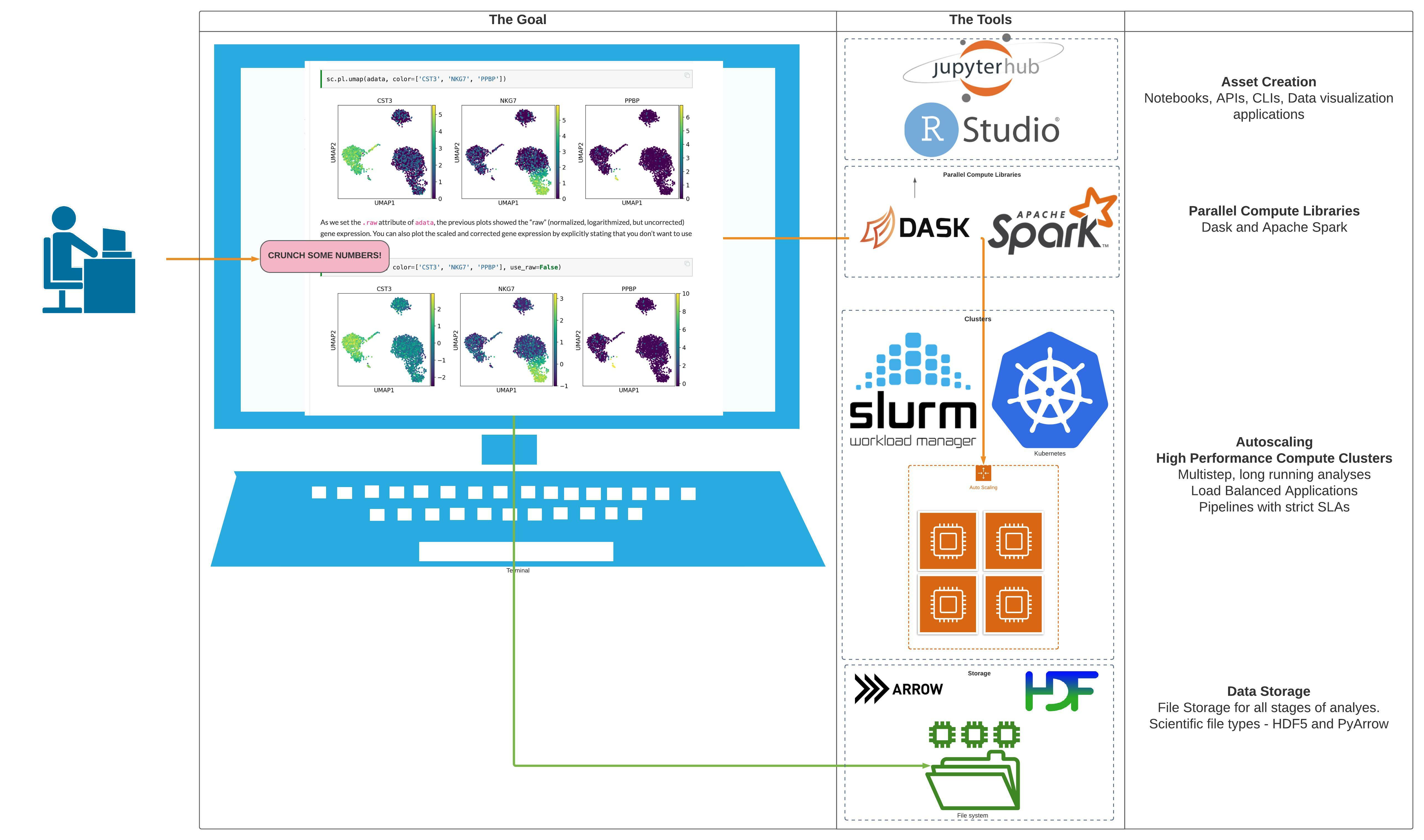
Terraform module to deploy a multiuser Jupyterhub + Dask Cluster on an existing EKS cluster using the DaskHub helm chart. It deploys the DaskHub Helm chart, and optionally configures SSL for you if you are using a domain name on AWS hosted with Route53.
This project is part of the "BioAnalyze" project, which aims to make High Performance Compute Architecture accessible to everyone.
It's 100% Open Source and licensed under the APACHE2.
This module installs the Bitnami Apache Airflow helm chart to an AWS EKS cluster. It will optionally setup an NGINX ingress for SSL and create AWS Route53 records.
IMPORTANT: We do not pin modules to versions in our examples because of the difficulty of keeping the versions in the documentation in sync with the latest released versions. We highly recommend that in your code you pin the version to the exact version you are using so that your infrastructure remains stable, and update versions in a systematic way so that they do not catch you by surprise.
Also, because of a bug in the Terraform registry (hashicorp/terraform#21417), the registry shows many of our inputs as required when in fact they are optional. The table below correctly indicates which inputs are required.
For a complete example, see examples/complete.
For automated tests of the complete example using bats and Terratest (which tests and deploys the example on AWS), see test.
resource "random_string" "computed_values" {
length = 10
special = false
lower = true
upper = false
override_special = ""
}
locals {
helm_release_merged_values_file = abspath("helm_charts/airflow/computed-${random_string.computed_values.result}-values.yaml")
}
# If the computed-values file does not exist we must create it first
resource "null_resource" "create_merged_file" {
triggers = {
always_run = timestamp()
}
provisioner "local-exec" {
command = <<EOT
touch ${local.helm_release_merged_values_file}
EOT
}
}
module "helm_release_airflow" {
depends_on = [
null_resource.create_merged_file,
]
source = "dabble-of-devops-bioanalyze/eks-bitnami-apache-airflow/aws"
helm_release_name = var.airflow_release_name
helm_release_values_dir = abspath(var.airflow_helm_values_dir)
# this can be named whatever you like, including values.yaml, just make sure that the file exists before starting up the module
helm_release_merged_values_file = local.helm_release_merged_values_file
enable_ssl = true
letsencrypt_email = "hello@gmail.com"
# This must be a domain name registered with AWS
# And it must have a wildcard ACM
# The module will configure SSL and at the end you will have
# https://airflow.example.com
aws_route53_zone_name = "example.com."
aws_route53_record_name = "airflow"
context = module.this.context
}
output "helm_release_airflow" {
value = module.helm_release_airflow
}Here is an example of using this module:
examples/complete- complete example of using this module
Available targets:
help Help screen
help/all Display help for all targets
help/short This help short screen
lint Lint terraform code
| Name | Version |
|---|---|
| terraform | >= 0.13 |
| http | >= 2.1.0 |
| local | >= 1.2 |
| random | >= 2.2 |
| Name | Version |
|---|---|
| aws | n/a |
| kubernetes | n/a |
| local | >= 1.2 |
| null | n/a |
| template | n/a |
| Name | Source | Version |
|---|---|---|
| airflow | git::https://github.com/dabble-of-devops-bioanalyze/terraform-aws-eks-helm.git | n/a |
| s3_bucket | cloudposse/s3-bucket/aws | n/a |
| this | cloudposse/label/null | 0.25.0 |
| Name | Type |
|---|---|
| aws_iam_policy.s3-logs-eks | resource |
| aws_iam_role.logs | resource |
| kubernetes_namespace.airflow_ns | resource |
| kubernetes_service_account.eks_s3 | resource |
| local_file.rendered_airflow_db | resource |
| local_file.rendered_airflow_remote_logs | resource |
| local_file.rendered_auth | resource |
| null_resource.fernet | resource |
| null_resource.helm_dirs | resource |
| null_resource.sleep_airflow_update | resource |
| aws_caller_identity.current | data source |
| aws_elb.airflow | data source |
| aws_iam_policy_document.assume_role_with_oidc | data source |
| aws_iam_policy_document.s3-logs-eks | data source |
| aws_partition.current | data source |
| kubernetes_service.airflow | data source |
| local_file.fernet | data source |
| template_file.airflow_external_db | data source |
| template_file.airflow_remote_logging | data source |
| template_file.auth | data source |
| Name | Description | Type | Default | Required |
|---|---|---|---|---|
| additional_tag_map | Additional key-value pairs to add to each map in tags_as_list_of_maps. Not added to tags or id.This is for some rare cases where resources want additional configuration of tags and therefore take a list of maps with tag key, value, and additional configuration. |
map(string) |
{} |
no |
| airflow_password | Initial airflow password | string |
"Password#123" |
no |
| airflow_user | Initial airflow user | string |
"user" |
no |
| attributes | ID element. Additional attributes (e.g. workers or cluster) to add to id,in the order they appear in the list. New attributes are appended to the end of the list. The elements of the list are joined by the delimiterand treated as a single ID element. |
list(string) |
[] |
no |
| aws_elb_dns_name | DNS of AWS ELB for ingress controller | string |
n/a | yes |
| aws_elb_zone_id | AWS ELB zone id for ingress controller | string |
n/a | yes |
| aws_route53_record_name | Record name to add to aws_route_53. Must be a valid subdomain - www,app,etc | string |
"www" |
no |
| aws_route53_zone_id | AWS route_53 zone ID for airflow | string |
"" |
no |
| aws_route53_zone_name | Name of the zone to add records. Do not forget the trailing '.' - 'test.com.' | string |
"bioanalyzedev.com." |
no |
| context | Single object for setting entire context at once. See description of individual variables for details. Leave string and numeric variables as null to use default value.Individual variable settings (non-null) override settings in context object, except for attributes, tags, and additional_tag_map, which are merged. |
any |
{ |
no |
| delimiter | Delimiter to be used between ID elements. Defaults to - (hyphen). Set to "" to use no delimiter at all. |
string |
null |
no |
| descriptor_formats | Describe additional descriptors to be output in the descriptors output map.Map of maps. Keys are names of descriptors. Values are maps of the form {<br> format = string<br> labels = list(string)<br>}(Type is any so the map values can later be enhanced to provide additional options.)format is a Terraform format string to be passed to the format() function.labels is a list of labels, in order, to pass to format() function.Label values will be normalized before being passed to format() so they will beidentical to how they appear in id.Default is {} (descriptors output will be empty). |
any |
{} |
no |
| enable_ssl | Enable SSL Support? | bool |
true |
no |
| enabled | Set to false to prevent the module from creating any resources | bool |
null |
no |
| encrypt_s3_logs | n/a | string |
"False" |
no |
| environment | ID element. Usually used for region e.g. 'uw2', 'us-west-2', OR role 'prod', 'staging', 'dev', 'UAT' | string |
null |
no |
| external_db_host | n/a | string |
"" |
no |
| external_db_name | n/a | string |
"airflow" |
no |
| external_db_port | n/a | number |
5432 |
no |
| external_db_secret | n/a | string |
"" |
no |
| external_db_type | n/a | string |
"postgres" |
no |
| external_db_user | n/a | string |
"" |
no |
| helm_release_chart | helm release chart | string |
"airflow" |
no |
| helm_release_name | helm release name | string |
"airflow" |
no |
| helm_release_namespace | helm release namespace for BioAnalyze deployment | string |
n/a | yes |
| helm_release_repository | helm release chart repository | string |
"https://charts.bitnami.com/bitnami" |
no |
| helm_release_values_dir | Directory to put rendered values template files or additional keys. Should be helm_charts/{helm_release_name} | string |
"helm_charts" |
no |
| helm_release_values_files | helm release values files - paths values files to add to helm install --values {} | list(string) |
[] |
no |
| helm_release_values_service_port | Service port to set for exposing the airflow service | string |
"80" |
no |
| helm_release_values_service_type | Service type to set for exposing the airflow service. The default is to use the ClusterIP and an ingress. Alternative is to use a LoadBalancer, but this only recommended for testing. | string |
"ClusterIP" |
no |
| helm_release_version | helm release version | string |
"12.0.10" |
no |
| helm_release_wait | n/a | bool |
false |
no |
| id_length_limit | Limit id to this many characters (minimum 6).Set to 0 for unlimited length.Set to null for keep the existing setting, which defaults to 0.Does not affect id_full. |
number |
null |
no |
| label_key_case | Controls the letter case of the tags keys (label names) for tags generated by this module.Does not affect keys of tags passed in via the tags input.Possible values: lower, title, upper.Default value: title. |
string |
null |
no |
| label_order | The order in which the labels (ID elements) appear in the id.Defaults to ["namespace", "environment", "stage", "name", "attributes"]. You can omit any of the 6 labels ("tenant" is the 6th), but at least one must be present. |
list(string) |
null |
no |
| label_value_case | Controls the letter case of ID elements (labels) as included in id,set as tag values, and output by this module individually. Does not affect values of tags passed in via the tags input.Possible values: lower, title, upper and none (no transformation).Set this to title and set delimiter to "" to yield Pascal Case IDs.Default value: lower. |
string |
null |
no |
| labels_as_tags | Set of labels (ID elements) to include as tags in the tags output.Default is to include all labels. Tags with empty values will not be included in the tags output.Set to [] to suppress all generated tags.Notes: The value of the name tag, if included, will be the id, not the name.Unlike other null-label inputs, the initial setting of labels_as_tags cannot bechanged in later chained modules. Attempts to change it will be silently ignored. |
set(string) |
[ |
no |
| letsencrypt_email | Email to use for https setup. Not needed unless enable_ssl | string |
"hello@gmail.com" |
no |
| logging_level | n/a | string |
"DEBUG" |
no |
| name | ID element. Usually the component or solution name, e.g. 'app' or 'jenkins'. This is the only ID element not also included as a tag.The "name" tag is set to the full id string. There is no tag with the value of the name input. |
string |
null |
no |
| namespace | ID element. Usually an abbreviation of your organization name, e.g. 'eg' or 'cp', to help ensure generated IDs are globally unique | string |
null |
no |
| oidc_fully_qualified_subjects | The fully qualified OIDC subjects to be added to the role policy | set(string) |
[ |
no |
| provider_urls | List of URLs of the OIDC Providers | list |
[] |
no |
| regex_replace_chars | Terraform regular expression (regex) string. Characters matching the regex will be removed from the ID elements. If not set, "/[^a-zA-Z0-9-]/" is used to remove all characters other than hyphens, letters and digits. |
string |
null |
no |
| region | AWS Region | string |
"us-east-1" |
no |
| remote_base_log_folder | n/a | string |
"" |
no |
| remote_log_conn_id | n/a | string |
"aws_default" |
no |
| remote_logging | n/a | bool |
false |
no |
| render_cluster_issuer | n/a | bool |
false |
no |
| render_ingress | n/a | bool |
true |
no |
| stage | ID element. Usually used to indicate role, e.g. 'prod', 'staging', 'source', 'build', 'test', 'deploy', 'release' | string |
null |
no |
| tags | Additional tags (e.g. {'BusinessUnit': 'XYZ'}).Neither the tag keys nor the tag values will be modified by this module. |
map(string) |
{} |
no |
| tenant | ID element _(Rarely used, not included by default)_. A customer identifier, indicating who this instance of a resource is for | string |
null |
no |
| use_existing_ingress | n/a | bool |
true |
no |
| use_external_db | n/a | bool |
false |
no |
| Name | Description |
|---|---|
| aws_elb_airflow | n/a |
| helm_notes | n/a |
| helm_release | n/a |
Like this project? Please give it a ★ on our GitHub! (it helps a lot)
Check out these related projects.
- terraform-aws-eks-autoscaling - Terraform module to provision an Autoscaling EKS Cluster. Acts as a wrapper around cloudposse/terraform-aws-eks-cluster and cloudposse/terraform-aws-eks-node-groups
- terraform-aws-eks-cluster - Terraform module to deploy an AWS EKS Cluster.
- terraform-aws-eks-node-group - Terraform module to provision an EKS Node Group
- Zero to Kubernetes - JupyterHub allows users to interact with a computing environment through a webpage. As most devices have access to a web browser, JupyterHub makes it is easy to provide and standardize the computing environment for a group of people (e.g., for a class of students or an analytics team).
This project will help you set up your own JupyterHub on a cloud/on-prem k8s environment and leverage its scalable nature to support a large group of users. Thanks to Kubernetes, we are not tied to a specific cloud provider.
-
Dask Gateway - Dask Gateway provides a secure, multi-tenant server for managing Dask clusters. It allows users to launch and use Dask clusters in a shared, centrally managed cluster environment, without requiring users to have direct access to the underlying cluster backend (e.g. Kubernetes, Hadoop/YARN, HPC Job queues, etc…).
-
DaskHub - This chart provides a multi-user, Dask-Gateway enabled JupyterHub. It combines the JupyterHub and Dask Gateway helm charts.
-
Dask - Dask is a flexible library for parallel computing in Python.
-
terraform-aws-ec2-autoscale-group - Terraform module to provision Auto Scaling Group and Launch Template on AWS
-
terraform-aws-ec2-instance - Terraform module for providing a general purpose EC2 instance
-
terraform-aws-ec2-instance-group - Terraform module for provisioning multiple general purpose EC2 hosts for stateful applications
-
terraform-null-label - Terraform module designed to generate consistent names and tags for resources. Use terraform-null-label to implement a strict naming convention.
For additional context, refer to some of these links.
- Bitnami Tutorial: Secure Kubernetes Services with Ingress, TLS and Let's Encrypt - Set up the NGINX Ingress Controller in our cluster and create Ingress routes for an example application. You'll learn how Ingress objects are defined, including how to configure TLS and basic authentication.
- Cert Manager Release Version Info -
- Cert Manager Installation with Helm -
- Terraform Standard Module Structure - HashiCorp's standard module structure is a file and directory layout we recommend for reusable modules distributed in separate repositories.
- Terraform Module Requirements - HashiCorp's guidance on all the requirements for publishing a module. Meeting the requirements for publishing a module is extremely easy.
- Terraform
random_integerResource - The resource random_integer generates random values from a given range, described by the min and max attributes of a given resource. - Terraform Version Pinning - The required_version setting can be used to constrain which versions of the Terraform CLI can be used with your configuration
Got a question? We got answers.
File a GitHub issue, send us an jillian@dabbleofdevops.com.
I'll help you build your data science cloud infrastructure from the ground up so you can own it using open source software. Then I'll show you how to operate it and stick around for as long as you need us.
Work directly with me via email, slack, and video conferencing.
- Scientific Workflow Automation and Optimization. Got workflows that are giving you trouble? Let's work together to ensure that your analyses run with or without your scientists being fully caffeinated.
- High Performance Compute Infrastructure. Highly available, auto scaling clusters to analyze all the (bioinformatics related!) things. All setups are completely integrated with your workflow system of choice, whether that is Airflow, Prefect, Snakemake or Nextflow.
- Kubernetes and AWS Batch Setup for Apache Airflow Orchestrate your Bioinformatics Workflows with Apache Airflow. Get full auditing, SLA, logging and monitoring for your workflows running on AWS Batch.
- High Performance Compute Setup that Int You'll have built-in governance with accountability and audit logs for all changes.
- Docker Images Get advice and hands on training for your team to build complex software stacks onto docker images.
- Training. You'll receive hands-on training so your team can operate what we build.
- Questions. You'll have a direct line of communication between our teams via a Shared Slack channel.
- Troubleshooting. You'll get help to triage when things aren't working.
- Bug Fixes. We'll rapidly work with you to fix any bugs in our projects.
Please use the issue tracker to report any bugs or file feature requests.
If you are interested in being a contributor and want to get involved in developing this project or help out with other projects, I would love to hear from you! Shoot me an email at jillian@dabbleofdevops.com.
In general, PRs are welcome. We follow the typical "fork-and-pull" Git workflow.
- Fork the repo on GitHub
- Clone the project to your own machine
- Commit changes to your own branch
- Push your work back up to your fork
- Submit a Pull Request so that we can review your changes
The README.md is created using the standard CloudPosse template that has been modified to use BioAnalyze information and URLs, and other documentation is generated using jupyter-book.
Terraform code does not render properly when using the literalinclude directive, so instead we use pygmentize to render it to html which is included directly.
.. raw:: html
:file: ./_html/main.tf.html
NOTE: Be sure to merge the latest changes from "upstream" before making a pull request!
Copyright © 2020-2022 Dabble of DevOps, SCorp
See LICENSE for full details.
Licensed to the Apache Software Foundation (ASF) under one
or more contributor license agreements. See the NOTICE file
distributed with this work for additional information
regarding copyright ownership. The ASF licenses this file
to you under the Apache License, Version 2.0 (the
"License"); you may not use this file except in compliance
with the License. You may obtain a copy of the License at
https://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing,
software distributed under the License is distributed on an
"AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY
KIND, either express or implied. See the License for the
specific language governing permissions and limitations
under the License.
All other trademarks referenced herein are the property of their respective owners.
|
Jillian Rowe |
Learn more at Dabble of DevOps
BioAnalyze is and will always be open source. If you've found any of these resources helpful, please consider donating to the continued development of BioAnalyze.