An R package to download and merge labeled single-cell RNA-seq data from the PanglaoDB database into a Seurat object.
This package requires R version 4.0 or higher. If you are using an older version of R you will be prompted to upgrade when you try to install the package.
The official release of rPanglaoDB is available on CRAN. To install it from there, you can use the following command:
> install.packages('rPanglaoDB', dependencies = TRUE)
If you have remotes installed, you can install the latest stable version of rPanglaoDB package directly from GitHub:
> remotes::install_github('dosorio/rPanglaoDB')
| Code | Function |
|---|---|
| getMarkers | Return a data frame with the list of samples from the panglaoDB database exhibiting a pattern of expression for a set of molecular markers. |
| getSampleComposition | Return a data frame with the the cell-type content for each sample from the panglaoDB database. |
| getSampleList | Return a data frame with the list of samples available at the panglaoDB database. |
| getSamples | Download and return the expression matrix and annotations from the panglaoDB database in a Seurat object. |
As any other R package rPanglaoDB can be loaded using the library function as follows:
> library(rPanglaoDB)
To access the list of available samples deposited in the PanglaoDB database you may use the getSamplesList() function:
> samplesList <- getSampleList()
This function returns a data frame with 6 columns matching with the information provided here by the PanglaoDB database.
> head(samplesList)
SRA SRS Tissue Protocol Species Cells
1 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 6501
2 SRA570744 SRS2253536 Lung mesenchyme 10x chromium Mus musculus 4611
3 SRA598936 SRS2428405 Kidney cortex 10x chromium Homo sapiens 3759
4 SRA644036 SRS2808714 Cervical and lumbar spinal cord 10x chromium Mus musculus 1025
5 SRA670243 SRS3078084 Ventral midbrain 10x chromium Mus musculus 5603
6 SRA689041 SRS3166675 Colon 10x chromium Mus musculus 2878
To access the cell-type content for each sample from the panglaoDB database you may use the getSampleComposition function. This function returns the cell-type composition of the samples included in the PanglaoDB database in a data frame with 8 columns. For example, to retrieve the sample composition of the sample with SRS = SRS2119548 you may use the following code:
> scSRS2119548 <- getSampleComposition(srs = 'SRS2119548')
> head(scSRS2119548)
SRA SRS Tissue Protocol Species Cluster Cells Cell Type
1.1 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 0 1572 Unknown
1.2 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 1 563 Unknown
1.3 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 2 280 Unknown
1.4 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 3 270 Unknown
1.5 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 4 220 Unknown
1.6 SRA553822 SRS2119548 Cultured embryonic stem cells 10x chromium Homo sapiens 5 192 Unknown
Retrieved information match with the SRS2119548 reported record from the PanglaoDB available here.
To access the list of available samples with specific expression patterns you may use the getMarkers() function. This function returns the output of a query submitted through here in the PanglaoDB database.
As an example, below we show how to retrieve the list of clusters containing two specific types of Endothelial cells. This type of cells act as barriers between vessels and tissues (Aman et al., 2016). They are known to control the flow of substances and fluids into and out of a tissue. Endothelial cells line blood vessels and lymphatic vessels, and are found exclusively in vascularized tissue (Bautch and Caron, 2015). Endothelial cells can be classified on the basis of a set of marker genes, for example, Lymphatic Endothelial Cells (LEC) are PECAM and PDPN positive, meanwhile Blood Endothelial Cells (BEC) are PECAM1 and VWF positive but negative for PDPN and ACTA2.
> BEC <- getMarkers(include = c('PECAM1', 'VWF'), exclude = c('PDPN', 'ACTA2'))
> head(BEC)
SRA SRS Specie Tissue Cluster Cell-Type Markers
1 SRA646572 SRS2833946 Homo sapiens Human embryo forebrain 28 Endothelial cells +PECAM1+VWF-PDPN-ACTA2
2 SRA646572 SRS2833947 Homo sapiens Human embryo forebrain 24 Endothelial cells +PECAM1+VWF-PDPN-ACTA2
3 SRA594999 SRS2397417 Homo sapiens Umbilical vein endothelial cells 0 Endothelial cells +PECAM1+VWF-PDPN-ACTA2
4 SRA594999 SRS2397417 Homo sapiens Umbilical vein endothelial cells 2 Endothelial cells +PECAM1+VWF-PDPN-ACTA2
5 SRA594999 SRS2397417 Homo sapiens Umbilical vein endothelial cells 3 Endothelial cells +PECAM1+VWF-PDPN-ACTA2
6 SRA594999 SRS2397417 Homo sapiens Umbilical vein endothelial cells 4 Unknown +PECAM1+VWF-PDPN-ACTA2
> LEC <- getMarkers(include = c('PECAM1', 'PDPN', 'PROX1'))
> head(LEC)
SRA SRS Specie Tissue Cluster Cell-Type Markers
1 SRA640325 SRS2769051 Homo sapiens Lung proximal airway stromal cells 17 Endothelial cells +PECAM1+PDPN+PROX1
2 SRA703206 SRS3296613 Homo sapiens Colon (Ulcerative Colitis) 15 Unknown +PECAM1+PDPN+PROX1
3 SRA782908 SRS3815606 Homo sapiens Decidua 13 Endothelial cells +PECAM1+PDPN+PROX1
4 SRA637291 SRS2749416 Mus musculus Left Ventricle 17 Endothelial cells +PECAM1+PDPN+PROX1
5 SRA652149 SRS2862117 Mus musculus Lateral geniculate nucleus 11 Interneurons +PECAM1+PDPN+PROX1
6 SRA611634 SRS2532206 Mus musculus Lung 18 Endothelial cells +PECAM1+PDPN+PROX1
Once the desired samples to be downloaded are identified, the count matrices can be downloaded using the getSamples function. In the example below, we show how to download the set of Human Lymphatic Endothelial Cells applying two filters in the getSample function to the set of identified samples containing the desired phenotype (PECAM1+, PDPN+, PROX1+). By default, the output of the function is a Seurat object with all the samples merged. In this case is an object containing 1124 human endothelial cells.
> countsLEC <- getSamples(srs = unique(LEC$SRS), celltype = 'Endothelial cells', specie = 'Homo sapiens')
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100%
> countsLEC
An object of class Seurat
39551 features across 1124 samples within 1 assay
Active assay: RNA (39551 features, 0 variable features)
Metadata associated with the downloaded count matrices can be accessed using the [[]] operator.
> head(countsLEC[[]])
orig.ident nCount_RNA nFeature_RNA CellTypes panglaoCluster Tissue Specie
AAACCTGTCAGTACGT SRS2769051 3137 1526 Endothelial cells 17 Lung proximal airway stromal cells Homo sapiens
AAGGCAGAGGGAGTAA SRS2769051 1041 677 Endothelial cells 17 Lung proximal airway stromal cells Homo sapiens
ACCTTTAAGTAGGTGC SRS2769051 2431 1239 Endothelial cells 17 Lung proximal airway stromal cells Homo sapiens
ACGAGGAAGATGAGAG SRS2769051 2928 1470 Endothelial cells 17 Lung proximal airway stromal cells Homo sapiens
ACGGAGACAAGCTGTT SRS2769051 1971 1028 Endothelial cells 17 Lung proximal airway stromal cells Homo sapiens
AGACGTTGTGCCTTGG SRS2769051 1176 750 Endothelial cells 17 Lung proximal airway stromal cells Homo sapiens
Optionally if the unmerged samples are needed, you may set the merge parameter as FALSE. In this case the output is a list containing n number of Seurat objects as samples requested in the input.
> countsLEC <- getSamples(srs = unique(LEC$SRS), celltype = 'Endothelial cells', specie = 'Homo sapiens', merge = FALSE)
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100%
> countsLEC
$SRS2769051
An object of class Seurat
35225 features across 36 samples within 1 assay
Active assay: RNA (35225 features, 0 variable features)
$SRS3296613
An object of class Seurat
32131 features across 860 samples within 1 assay
Active assay: RNA (32131 features, 0 variable features)
$SRS3815606
An object of class Seurat
31724 features across 228 samples within 1 assay
Active assay: RNA (31724 features, 0 variable features)
Once downloaded and merged the desired samples, some postprocessing is required to identify the cells exhibiting the desired phenotype. For that purpose, here we show the process how to integrate all the samples using Seurat and Harmony. The cluster exhibiting the desired phenotype is identified using the Nebulosa package.
> set.seed(1)
> countsLEC <- Seurat::NormalizeData(countsLEC)
> countsLEC <- Seurat::FindVariableFeatures(countsLEC)
> countsLEC <- Seurat::ScaleData(countsLEC)
> countsLEC <- Seurat::RunPCA(countsLEC, verbose = FALSE)
> countsLEC <- harmony::RunHarmony(countsLEC, group.by.vars = 'orig.ident')
> countsLEC <- Seurat::FindNeighbors(countsLEC, reduction = 'harmony')
> countsLEC <- Seurat::FindClusters(countsLEC)
> countsLEC <- Seurat::RunTSNE(countsLEC, reduction = 'harmony')
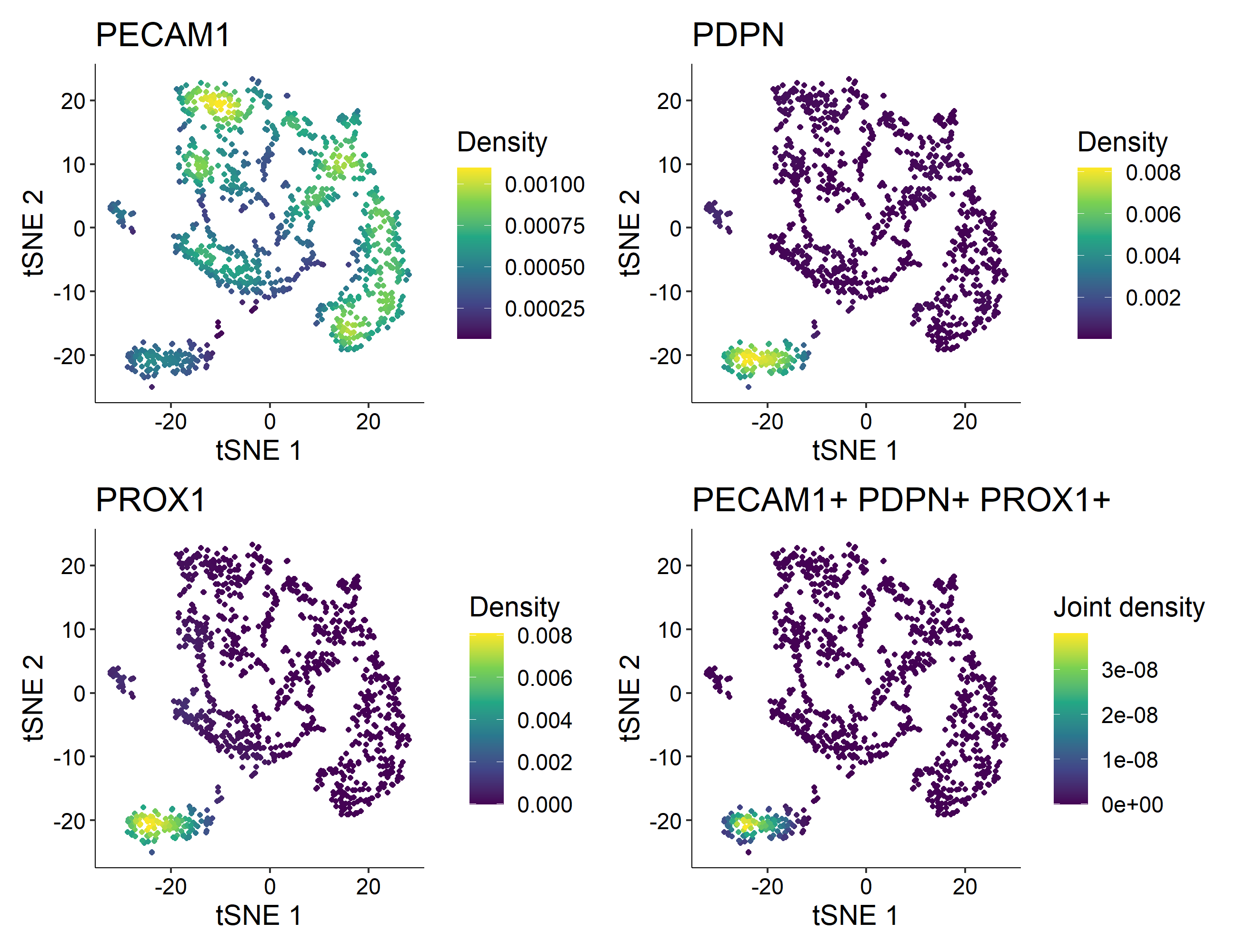
> Nebulosa::plot_density(countsLEC, features = c('PECAM1', 'PDPN', 'PROX1'), joint = TRUE)
In this example, cluster 4 is the one containing 121 Human Lymphatic Endothelial Cells with constitutive expression of PECAM1, PDPN, and PROX1.
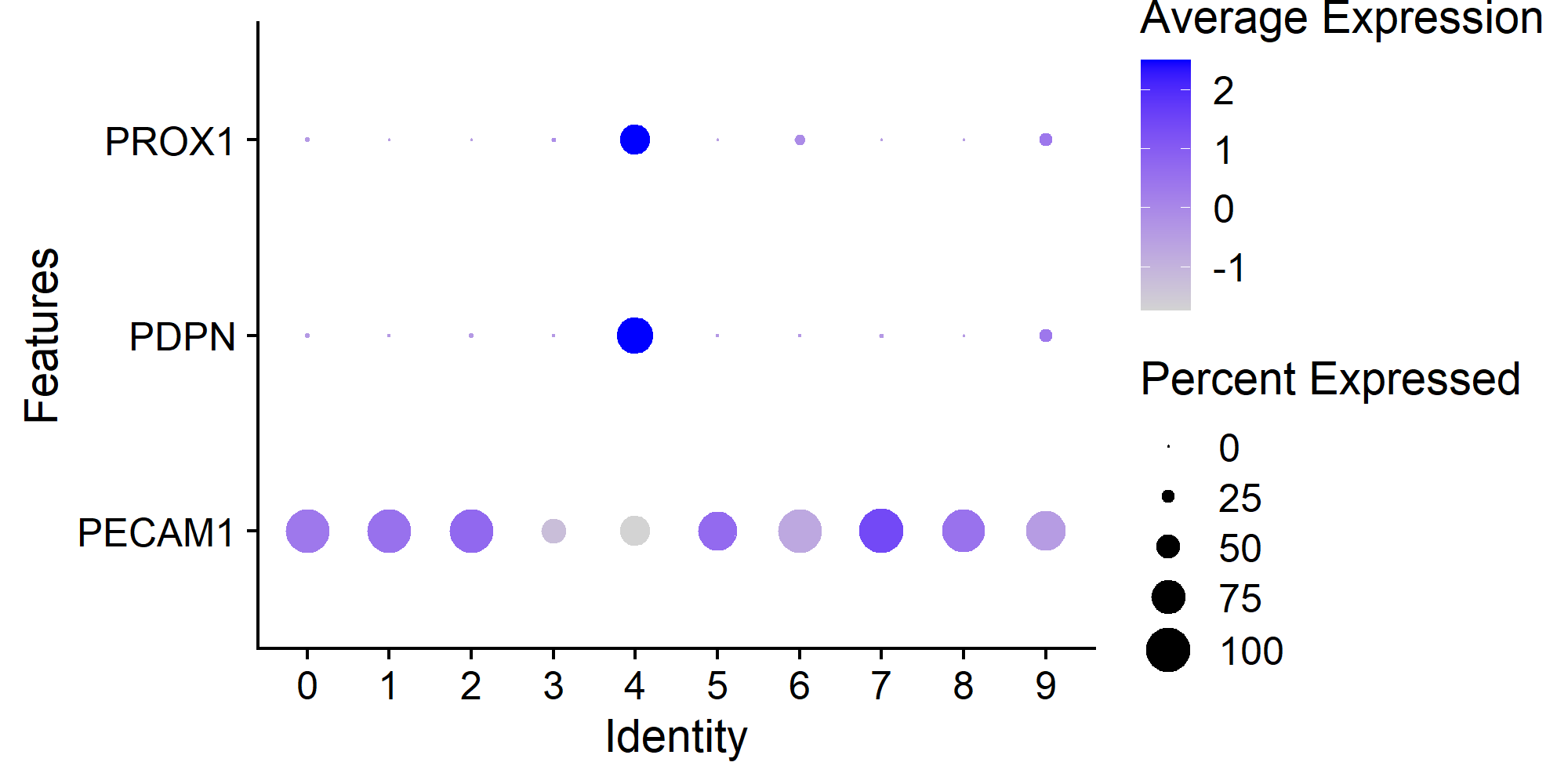
> Seurat::DotPlot(countsLEC, features = c('PECAM1', 'PDPN', 'PROX1')) + ggplot2::coord_flip()
> table(Seurat::Idents(countsLEC))
0 1 2 3 4 5 6 7 8 9
220 192 191 152 121 93 78 27 25 25
To show the potential of rPanglaoDB for collecting rare cell types by integrating multiple public datasets, we present a biological application collecting and characterizing a set of fibrocytes here
To cite package rPanglaoDB in publications use:
Daniel Osorio, Marieke Kuijjer and James J. Cai (2021). rPanglaoDB: Download and Merge Single-Cell RNA-Seq Data from the PanglaoDB Database. R package. https://CRAN.R-project.org/package=rPanglaoDB
A BibTeX entry for LaTeX users is
@Manual{,
title = {rPanglaoDB: Download and Merge Single-Cell RNA-Seq Data from the PanglaoDB Database},
author = {Daniel Osorio and Marieke Kuijjer and James J. Cai},
year = {2021},
note = {R package},
url = {https://CRAN.R-project.org/package=rPanglaoDB},
}