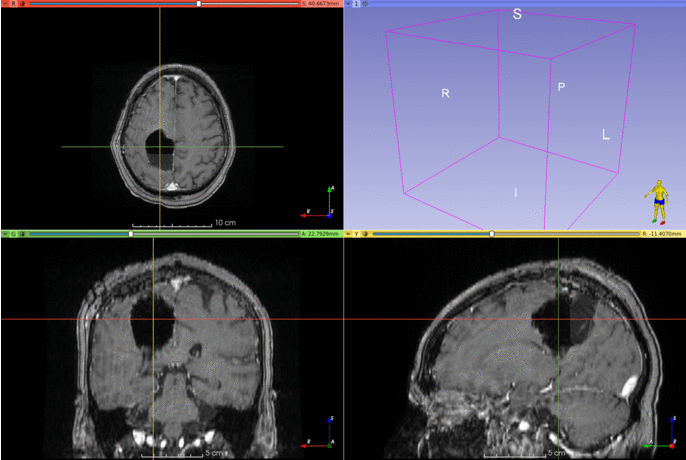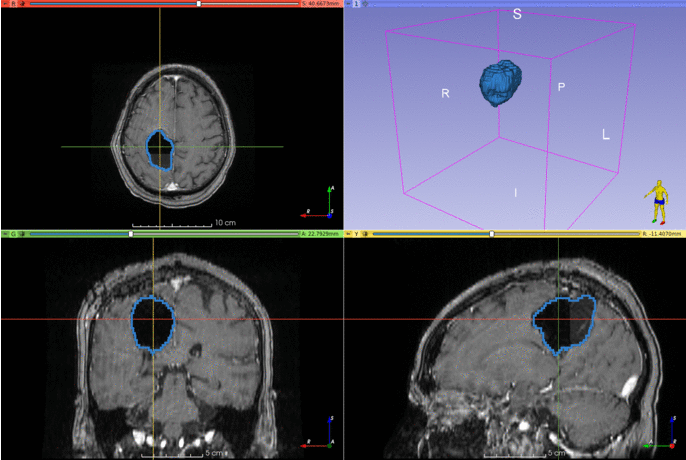
Automatic segmentation of postoperative brain resection cavities from magnetic resonance images (MRI) using a convolutional neural network (CNN) trained with PyTorch 1.7.1.
It's recommended to use conda.
A 6-GB GPU is large enough to segment an image in an MNI space of size 193 × 229 × 193.
conda create -n resseg python=3.8 -y
conda activate resseg
pip install light-the-torch
ltt install torch
pip install resseg
resseg --helpBelow are two examples of cavity segmentation for tumor and epilepsy surgery. The epilepsy example includes registration to the MNI space. Both examples can be run online using Google Colab:
Example using an image from the Brain Images of Tumors for Evaluation database (BITE).
BITE=`resseg-download bite`
resseg $BITE -o bite_seg.nii.gzExample using an image from the EPISURG dataset.
Segmentation works best when images are in the MNI space, so resseg includes a tool
for this purpose (requires `antspyx).
pip install antspyx
EPISURG=`resseg-download episurg`
resseg-mni $EPISURG -t episurg_to_mni.tfm
resseg $EPISURG -o episurg_seg.nii.gz -t episurg_to_mni.tfmThe trained model can be used without installing resseg, but you'll need to install unet first:
pip install unet==0.7.7Then, in Python:
import torch
repo = 'fepegar/resseg'
model_name = 'ressegnet'
model = torch.hub.load(repo, model_name, pretrained=True)There is an experimental graphical user interface (GUI) built on top of 3D Slicer.
Visit this repository for additional information and installation instructions.
A quantitative analysis of the resected structures can be performed using a brain parcellation computed using GIF (3.0) or FreeSurfer.
from resseg.parcellation import GIFParcellation, FreeSurferParcellation
parcellation_path = 't1_seg_gif.nii.gz'
cavity_seg_on_preop_path = 'cavity_on_preop.nii.gz'
parcellation = GIFParcellation(parcellation_path)I used a sphere near the hippocampus to simulate the resection cavity segmentation, and the GIF parcellation in the FPG dataset of TorchIO.
parcellation.print_percentage_of_resected_structures(cavity_seg_on_preop_path)Percentage of each resected structure:
100% of Left vessel
83% of Left Inf Lat Vent
59% of Left Amygdala
58% of Left Hippocampus
26% of Left PIns posterior insula
24% of Left PP planum polare
21% of Left Basal Forebrain
18% of Left Claustrum
16% of Left PHG parahippocampal gyrus
15% of Left Pallidum
15% of Left Ent entorhinal area
13% of Left FuG fusiform gyrus
13% of Left Temporal White Matter
11% of Left Putamen
10% of Left Insula White Matter
5% of Left ITG inferior temporal gyrus
5% of Left periventricular white matter
5% of Left Ventral DC
The resection volume is composed of:
30% is Left Temporal White Matter
12% is Left Hippocampus
10% is Left Insula White Matter
7% is Left FuG fusiform gyrus
6% is Left Amygdala
4% is Left ITG inferior temporal gyrus
4% is Left PP planum polare
3% is Left Putamen
3% is Left Claustrum
3% is Left PIns posterior insula
3% is Left PHG parahippocampal gyrus
2% is [Unkown label: 4]
1% is Left Ent entorhinal area
1% is Left Pallidum
1% is Left Inf Lat Vent
1% is Left Ventral DC
parcellation.plot_bars(cavity_seg_on_preop_path)parcellation.plot_pie(cavity_seg_on_preop_path)If you use this library for your research, please cite the following publications:
If you use the EPISURG dataset, which was used to train the model, please cite the following publication: