!!! UNDER DEVELOPMENT !!!
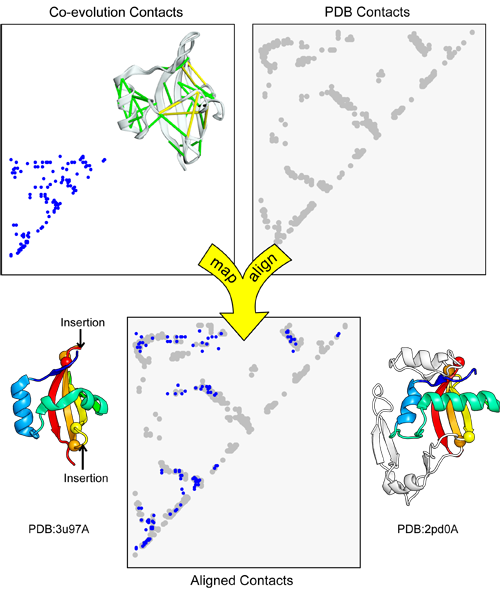
map_align takes two contact maps and returns an alignment that attempts to maximize the number of overlapping contacts while minimizing the number of gaps [1].
git clone https://github.com/gjoni/map_align
cd map_align
makeOptions: -s alignment.a3m - input, required
-c contacts.txt - input, required
-L list.txt with template IDs - input, required
-O prefix for saving top hits - output, optional
-N number of top hits to save 10
-T TM-score cleaning cut-off 0.80
-M max template size 1000
-I number of DP iterations 10
-t number of threads 1
-
-M MAXtemplates with more thanMAXresidues will be skipped (longer templates could take much more time to be aligned) -
-T TMtop N partial hits will be cleaned to exclude structurally similar matches: if two hits from the scan stage are similar with TM-score >TMthen only one (with the higher alignment score) will appear in the final pool -
-O PREFIXtop models will be saved as<PREFIX><TEMPLATE_ID>.pdband<PREFIX><TEMPLATE_ID>.aliwhereTEMPLATE_IDis an ID from thelistfile
List of templates is a text file with one entry per line. IDs of the templates are used in output and should not be longer than 10 characters.
/path/to/template1.pdb ID1
/path/to/template2.pdb ID2
...
Contact map is a list of residue-residue pairs in the following format (similar to CASP RR format):
i j d1 d2 p
i,j- indices of the two residues in contactd1,d2- distance limits defining a contact (currently not used)p- probability of the contact, should be in the range (0;1]
cd example
tar xf ecod70.tar.gz
Align a contact map to a library of templates (simplest call):
../map_align -s T0806.a3m -c T0806.con -L ecod70.list
Align a contact map to a library of templates saving top 5 hits at TM-score=70% identity cut-off and running the program on 4 cores (takes ~7 minutes on i5):
../map_align -s T0806.a3m -c T0806.con -L ecod70.list -N 5 -T 0.70 -t 4 -O T0806.
This package is a reimplementation of the original map_align program by S.Ovchinnikov [1] to allow for:
- direct use of PDB files as templates
- output of partial threads in PDB format
- cleaning of partial threads based on TM-score [2]
- multithreading
- kdtree library by John Tsiombikas
- C++ TM-align routine from Yang Zhang lab
[1] S Ovchinnikov et al. Protein Structure Determination using Metagenome sequence data. (2017) Science. 355(6322):294–8.
[2] Y Zhang & J Skolnick. TM-align: a protein structure alignment algorithm based on the TM-score. (2005) Nucleic Acids Res. 33(7):2302-9.