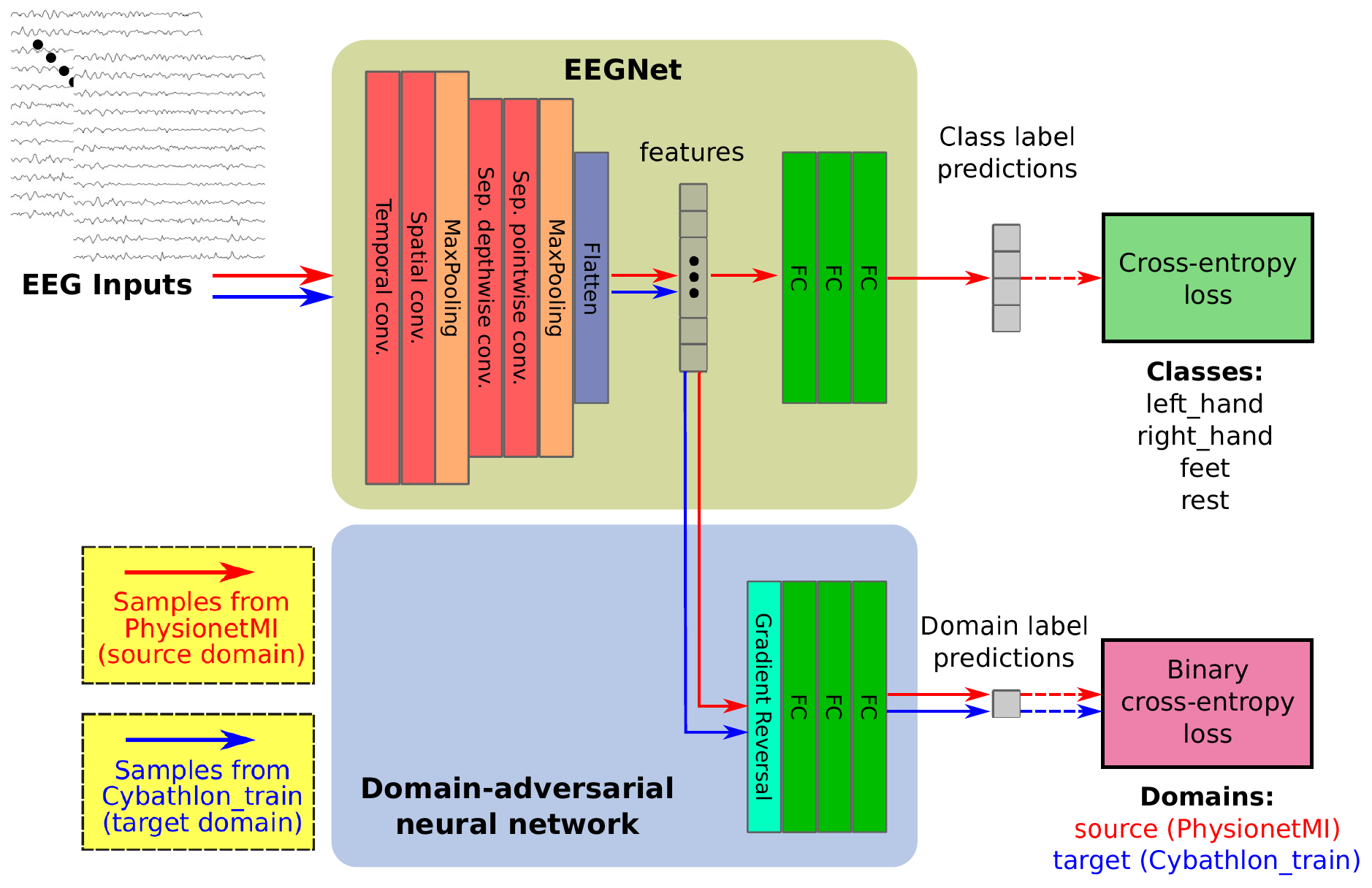
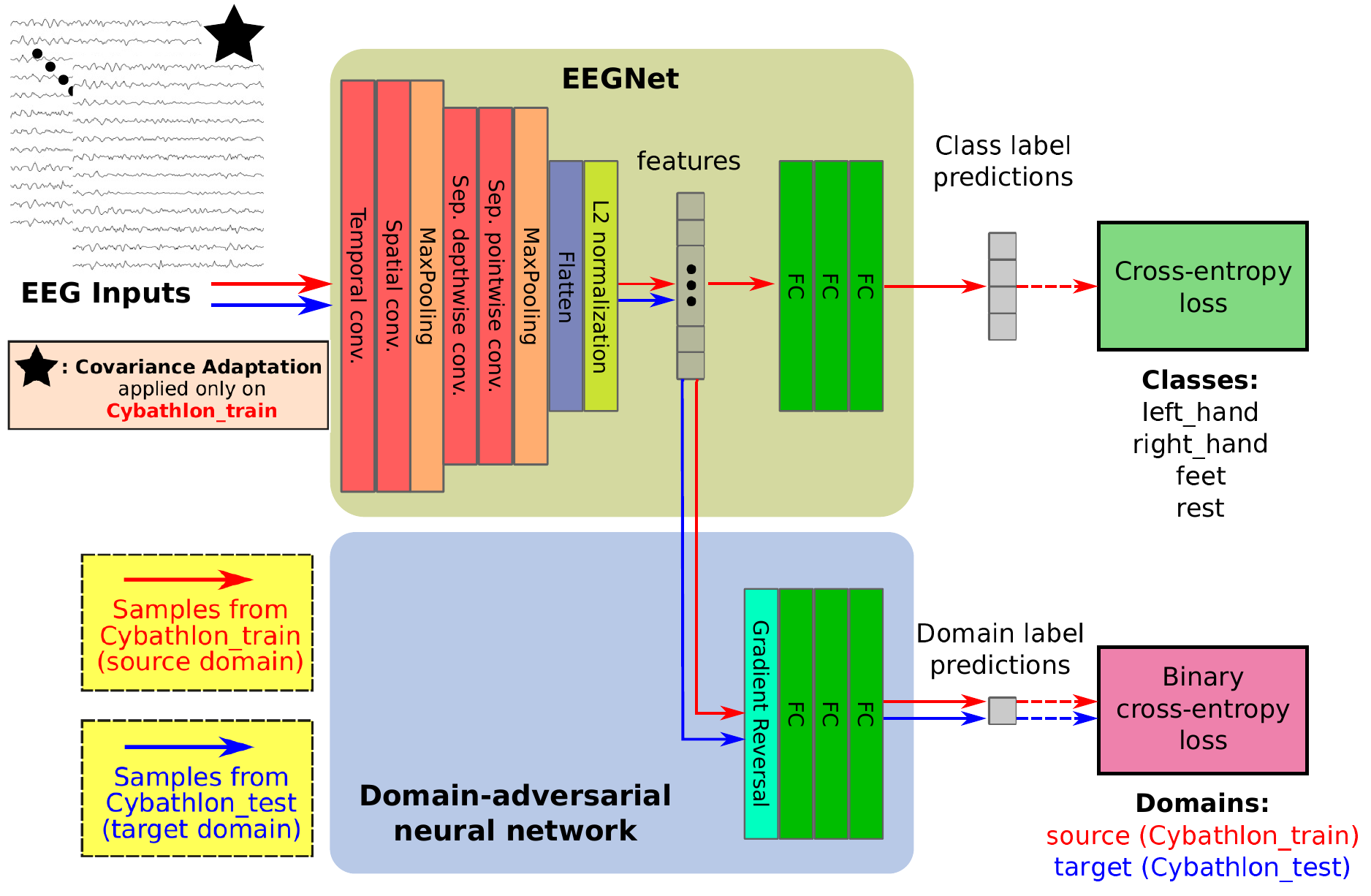
In this work, we present our method on the Motor Imagery classification task of the BEETL competition.
Our method includes two stages of training:
- 1st stage: Pre-training on PhysionetMI dataset + Unsupervised Domain Adaptation on Cybathlon dataset
- 2nd stage: Finetuning + Unsupervised Domain Adaptation + Covariance Adaptation on Cybathlon dataset
- We use the PhysionetMI dataset as our only external dataset, keeping the classes left_hand, right_hand, feet and rest.
- We use the BeetlMI Leaderboard dataset to experiment in the development phase
- We use the Cybathlon dataset to experiment in the final testing phase
All datasets should be downloaded at the directory ~/mne_data, with the following sub-directories:
~/mne_data/
├── MNE-eegbci-data/ (note: PhysionetMI)
├── MNE-beetlmileaderboard-data/ (note: BeetlMI)
└── MNE-beetlmitest-data/ (note: Cybathlon)
To download the Cybathlon dataset, you can execute the following commands
$ cd beetl
$ sh download_Cybathlon.sh Notes:
- We discard the EEG recording of subjects #104 and #106 on PhysionetMI, due to inconsistencies on the trial lengths.
- We keep the following 31 EEG electrodes, that exist in all of the datasets that we use:
'FC1', 'Pz', 'F3', 'CP6', 'P5', 'CP5', 'CP1', 'CP3', 'Fz', 'FC6', 'P2', 'P4', 'FC5', 'P3', 'C5', 'CP4', 'P7', 'C1', 'C3', 'C4', 'P8', 'C2', 'F4', 'CPz', 'C6', 'FC2', 'P1', 'Fp2', 'CP2', 'Fp1', 'P6'
EEG preprocessing has the following steps:
1) Bringing the EEG signals into the measurement unit of uV (microvolts)
2) Notch filtering to remove the 50Hz component, when necessary
3) Bandpass filtering in the range 4-38 Hz
4) Re-referencing the signals to the common average
5) Resampling signals to 100Hz
6) Adding two bipolar channels (specifically, the first bipolar channel is C3-C4 and the second bipolar channel is C4-C3). Thus, we have 33 EEG channels in total.
-
Motor imagery classification: The network architecture that is used for motor imagery classification, is a variant of the EEGNet implementation that exists in the braindecode toolbox. The original paper of EEGNet can be found here
-
Unsupervised Domain Adaptation: The network that is used to perform unsupervised domain adaptation, is based on the well-known DANN method, where a domain discriminator (classifier) is used, to predict whether a sample belongs to the source or target domain, combined with a Gradient Reversal Layer (GRL). The GRL implementation is the one that exists in the python-domain-adaptation toolbox of @jvanvugt.
We recommend installing the required packages using python's native virtual environment. For Python 3.4+, this can be done as follows:
$ python -m venv beetl_venv
$ source beetl_venv/bin/activate
(beetl_venv) $ pip install --upgrade pip
(beetl_venv) $ pip install -r requirements.txtTo train a model, you need to use src/run_exp_train.py, running the following command:
python src/run_exp_train.pyYou can inspect the various arguments that are passed to src/train.py.
By default, the src/run_exp_train.py script performs:
- Training on PhysionetMI dataset, as a first step that is later followed by finetuning on Cybathlon dataset. Batch size is set to
64and the training will be conducted for100epochs. - Unsupervised Domain Adaptation (UDA) on Cybathlon dataset. UDA is done with an increasing contribution of the domain classification loss to the total optimization loss, from
0.0to0.3, as the training process progresses. For details on the domain loss coefficient, check the original DANN paper, and the corresponding code atsrc/train.py.
The typical cross-entropy loss is used as the criterion for motor imagery classification, where the targets are the 4 aforementioned classes of PhysionetMI, by undersampling occasionaly as needed, to keep the dataset balanced with respect to sample occurence per class.
SGD is selected as the optimizer (momentum=0.9, weight decay=5e-4), with a warmup period of 20 epochs where the learning rate is increased linearly from 1e-5 to 0.01. After the initial warmup period, a cosine annealing scheduler is used to decrease the learning rate for the remaining 80 epochs. For this 1st stage, we use dropout with probability equal to 0.1 in EEGNet.
The Cybathlon training (labelled) set is used as the validation set of this stage. We can do that, as the labels of Cybathlon's training set have not been used yet, thus we get an idea of how our model generalizes on Cybathlon. The accuracy on this validation set is used to determine the best model, and save its weights in a checkpoint.
The training process will create the following directories:
json (note: contains the arguments of each experiment, stored in JSON format)
checkpoints (note: contains the saved model checkpoints, stored as .pth files)
preds (note: contains the predictions on a dataset, as a .txt file and as a .npy file)
To finetune a model, you need to use src/run_exp_finetune.py, running the following command:
python src/run_exp_finetune.pyYou can inspect the various arguments that are passed to src/train.py.
By default, the src/run_exp_finetune.py script performs:
- Finetuning on Cybathlon dataset, specifically on its labelled (training) set.
- UDA on Cybathlon dataset, specifically on its unlabelled (testing) set. UDA is done with a fixed contribution of the domain classification loss to the total optimization loss, as the domain loss coefficient is kept equal to
1.0. - Covariance Adaptation (CA) on Cybathlon dataset, specifically on its labelled (training) set. CA adapts the covariance matrix of each participant in the training set (i.e. the Cybathlon labelled set in this case), taking into consideration the covariance matrix of the same participant on the test set (i.e. the Cybathlon unlabelled set in this case). This is performed in the
braindecode/datasets/custom_dataset.pyscript. Covariance Adaptation is enabled by setting thebatch_alignargument toTrue(andpre_alignargument toFalse). We set the covariance mixing coefficient equal to0.5.
A pretrained model's checkpoint is loaded on EEGNet. The domain discriminator network is randomly initialized.
Batch size is set to 64 and finetuning is conducted for 130 epochs. The typical cross-entropy loss is used as the criterion for motor imagery classification, where the targets are the 4 aforementioned classes. No undersampling is applied in this stage.
AdamW is selected as the optimizer (weight decay=5e-4), using its PyTorch implementation with a constant learning rate of 1e-4 (no warmup period, no scheduler). For more details on AdamW, you can check the original paper here.
For this 2nd stage, we use dropout with probability equal to 0.3 in EEGNet. We freeze the parameters of the first convolutional layer (temporal convolution) of EEGNet. We also insert an L2 normalization layer, between the last convolution layer and the MLP classifier of EEGNet during the 2nd stage.
The BeetlMI training (labelled) set is used as the validation set of this stage. We can do that, as the labels of BeetlMI's training set have not been used yet. We get an idea of how our model generalizes on BeetlMI, which is somehow similar to Cybathlon. We performed a visual inspection of the Riemannian distances between the covariance matrices for all the participants of BeetlMI and Cybathlon, labelled and unlabelled sets. The distance matrix can be found in image format, at the file plots/cov_distances_BeetlMI_Cybathlon.png. We decided to discard participant #2 from the BeetlMI training set, when using it for validation purposes to assume the generalization on Cybathlon, as it seemed to have the most dissimilar statistics. The accuracy on this validation set is used to determine the best model, and save its weights in a checkpoint. This checkpoint is used to obtain the predictions on Cybathlon test (unlabelled) set. Please note that to conform with the target classes as needed to evaluate a submission on the BEETL competition (i.e., merging the classes "feet" and "rest" into one class named "other"), we replace every predicted label of class ID 3 (i.e. 4th class using zero-indexing), to class ID 2: other (0: left_hand, 1: right_hand).
The models checkpoints are as follows:
./checkpoints/
├── net_best_pretrained.pth
└── net_best_finetuned.pth
- Checkpoint after the 1st (pretraining) stage:
checkpoints/net_best_pretrained.pth - Checkpoint after the 2nd (finetuning) stage:
checkpoints/net_best_finetuned.pth
To evaluate a model, you need to use src/run_eval_pretrained.py or src/run_eval_finetuned.py, running the following commands:
python src/run_eval_pretrained.pyor
python src/run_eval_finetuned.pyYou can inspect the various arguments that are passed to src/eval.py.
The src/run_eval_pretrained.py script performs evaluation on Cybathlon dataset, specifically on its labelled (training) set. The obtained accuracy is ~40-41% on the 4-class problem, i.e. when not merging the classes "feet" and "rest" into one class named "other". The checkpoint used for this, is available at the file checkpoints/net_best_pretrained.pth.
The src/run_eval_finetuned.py script performs evaluation on Cybathlon dataset, specifically on its unlabelled (test) set. The obtained accuracy, when submitting the results on CodaLab's page for the final scoring phase of motor imagery, is ~55.7% on the 3-class problem, i.e. when merging the classes "feet" and "rest" into one class named "other". The checkpoint used for this, is available at the file checkpoints/net_best_finetuned.pth. Please note that the accuracy printed on the terminal when running this script, does not have any meaning, as we load a vector full of zeros to be used as the groundtruth labels of Cybathlon's test set.
The research of Georgios Zoumpourlis was supported by QMUL Principal's Studentship.
The current GitHub repo contains code parts from the following repositories (sometimes heavily chopped, keeping only the necessary tools). Credits go to their owners/developers. Their licenses are included in the corresponding folders.
Code in beetl: https://github.com/XiaoxiWei/NeurIPS_BEETL
Code in beetl: https://github.com/sylvchev/beetl-competition
Code in moabb: https://github.com/NeuroTechX/moabb
Code in braindecode: https://github.com/braindecode/braindecode
Code in skorch: https://github.com/skorch-dev/skorch
Code in braindecode/models/dann.py: https://github.com/jvanvugt/pytorch-domain-adaptation