-
Notifications
You must be signed in to change notification settings - Fork 210
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Can plotnine make a matrix of scatterplots? #16
Comments
|
You cannot do that with plotnine, though it is a goal of the project that a third party library should be able to implement such compound plots using plotnine. However, you can do it with seaborn. |
|
Sorry for reviving this post Is this answer still valid in 2021? |
|
Still nothing 'out of the box' I believe. Here's a start though: |
|
I was annoyed that the seaborn implementation of a pairplot did not allow me to change the shape of the point, only the color. This implementation in plotnine can do both shape and color. Note that I use |
|
@has2k1 Is there a third-party plotnine package where this function be sent to? |
It looks like with seaborn you can change the markers in pairplot:
|
|
@PaulHiemstra, I don't think there is. Please open a separate issue to track this. |
The issue here is that the colors and the markers are based on the same variable, this is not what I was looking for. I want to base the color on one variable, and shape on the other. the case of my visualisation this enables me to inspect the difference between the fitted clusters of a Kmeans and the underlying Species of Iris. |
I'll do that. |


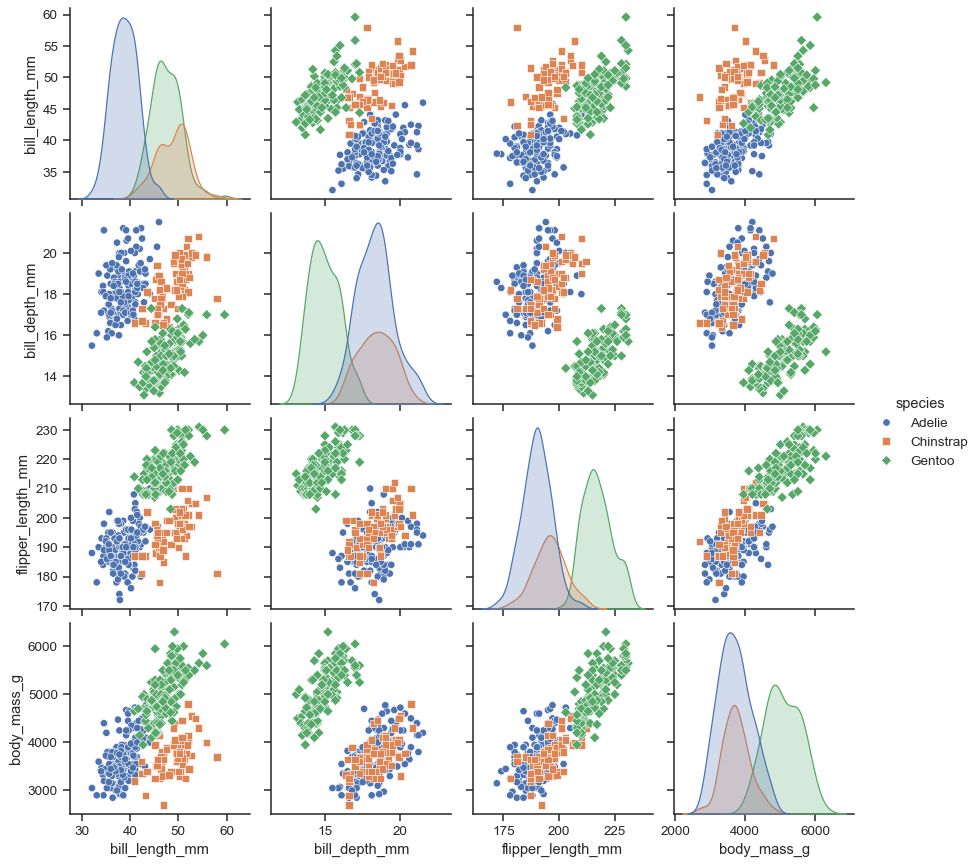
Something similar to R's
pairsorplotmatrixfunctions; see e.g. here?The text was updated successfully, but these errors were encountered: