Cinder is a highly modular spiking neuron simulator written in C++. In contrast to network simulators, it is optimised for simulating single neurons, allowing to analyse their behaviour, e.g. under changing parameters or varying input. Cinder is designed for expandability. Adding new membrane or synapse models is mainly a matter of writing down their differential equation in C++.
Spiking neuron models are generally given as a set of differential equations which usually cannot be solved in closed form. Unfortunately, this implies that analysing the behaviour of these models is – in most cases – restricted to numerical simulation of the neuron dynamics.
Cinder is a modular library which implements numerical simulation of single neurons, allowing to analyse the properties of the neuron models. In contrast to network simulators Cinder gives full control over the simulation process. It allows to record any neuron state variable, to use any number of current sources (such as synapses) and to use multiple different integrators.
Cinder requires a C++14 compatible compiler such as GCC 5 or clang 3.6,
as well as cmake 3.2. You can build and install Cinder using the following
commands:
git clone --depth 1 https://github.com/hbp-sanncs/cinder && cd cinder
mkdir build && cd build
cmake .. && make && make test
sudo make installNote that CMake will automatically download and build Google Test for the integrated integration and unit test suite.
Basically you only need to make sure that the cinder folder within the
cinder repository is in the search path of your C++ compiler, as done by
calling sudo make install in the above script. When using cmake for
your project you can simply include Cinder using
find_package(Cinder REQUIRED)
include_directories(${CINDER_INCLUDE_DIRS})If possible, you should use the -ffast-math -fno-finite-math-only compiler
flags for a considerable performance boost. Also note that with disabled
optimisation (-O0) Cinder is unusably slow. Enabling optimisation will give
you a 100 to 1000 times speedup.
Note that Cinder allows you to use either single or double precision floating
point numbers by setting the CINDER_REAL_WIDTH macro to either "4" or "8".
Usually, using double precision (default) floating point numbers has close to no
negative impact on performance. The floating point type used internally by
Cinder is called Real.
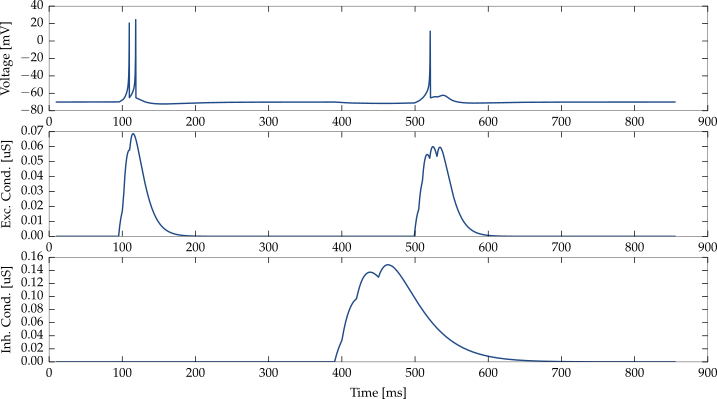
The following code simulates an Izhikevich neuron with two conductance based synapses – one excitatory synapse and one inhibitory synapse. The excitatory synapse receives spikes at about 100 and 500 ms, the second synapse at about 400 ms into the simulation.
#include <iostream>
#include <cinder/cinder.hpp>
using namespace cinder;
int main()
{
// Use the DormandPrince integrator with default target error
DormandPrinceIntegrator integrator;
// Record the neuron state as CSV to cout
CSVRecorder recorder(std::cout);
// Use the AutoController class to automatically abort the simulation
// once the neuron has settled to its resting state
AutoController controller;
// Assemble current source (here: two synapses)
auto current_source = make_current_source(
CondAlpha(20_nS, 10_ms, 30_mV, {95_ms, 100_ms, 102_ms, 110_ms, 499_ms,
505_ms, 510_ms, 520_ms, 530_ms}),
CondAlpha(50_nS, 30_ms, -80_mV, {390_ms, 400_ms, 420_ms, 450_ms}));
// Assemble the neuron (an Izhikevich neuron with default parameters)
auto neuron = make_neuron<Izhikevich>(current_source, [] (Time) {
// Lambda called whenever the neuron spikes
});
// Simulate the resulting ODE -- the recorder will record the results
make_solver(neuron, integrator, recorder, controller).solve();
}Plotting the resulting membrane potential and conductivity traces results in the following image:
Cinder is implemented as a C++ header-only template library. This allows the compiler to construct a customised code-path for exactly the simulation that is being performed, while keeping the actual model implementations extremely concise. With enabled optimisation, a single neuron simulation is converted into an inline blob of vectorised assembly without any external function calls. This way, Cinder is magnitudes faster than full network simulators simulating a single neuron.
Conceptually, each single neuron simulation is represented by an autonomous ordinary differential equation (ODE) which is automatically assembled at compile time using variadic templates. This single ODE representing the entire neuron, including its membrane and current sources, is then solved using a numerical differential equation integrator.
- LIF: Linear Integrate and Fire
- AdEx: Exponential Integrate and Fire Neuron with adaptive threshold
- Izhikevich: Quadratic Integrate and Fire Neuron
- MAT2: Multi-timescale Adaptive Threshold Integrade and Fire neuron with L=2 time constants
- HodgkinHuxley: Hodgkin-Huxley type model with Traub channel dynamics (same model as implemented in NEST as
hh_cond_exp_traub)
New neuron models can be easily implemented by simply defining the parameter and state vector and the corresponding differential equation. More neuron models will be added in the future.
- CondAlpha: Conductance-based synapse with alpha-function shaped decay
- CondExp: Conductance-based synapse with exponential-function shaped decay
- CurAlpha: Current-based synapse with alpha-function shaped decay
- CurExp: Current-based synapse with exponential-function shaped decay
- Delta: Dirac-Delta current pulse causing a defined voltage step
Furthermore, arbitrary current sources can be used to inject currents into the neuron. Pre-defined current sources include:
- ConstantCurrentSource: Current source which injects a constant current into the neuron
- StepCurrentSource: Current source which injects a constant current during a pre-defined time span into the neuron
- EulerIntegrator: The most simple differential equation integrator
- MidpointIntegrator: Second-order Runge-Kutta integrator
- RungeKuttaIntegrator: Fourth-order Runge-Kutta integrator
- DormandPrinceIntegrator: Fourth-order Runge-Kutta-based adaptive stepsize integrator
This project has been initiated by Andreas Stöckel and Christoph Jenzen in 2016 while working at Bielefeld University in the Cognitronics and Sensor Systems Group which is part of the Human Brain Project, SP 9.
This project and all its files are licensed under the GPL version 3 unless explicitly stated differently.