Third part softwares needed:
-Git: https://git-scm.com/downloads
-Anaconda3: https://www.anaconda.com/products/individual
-PyMOL: https://pymol.org/2/#download
Protein Contact Networks Miner is a command line tool (and now a Graphic User Interface) designed for annotate allosteric domains of a protein based of his rappresentation trough an unweighted graph, this graph is also called Protein Contact Network.
A Protein Contact Network is an unweighted graph where: nodes are the amino acids of the protein and exists an edge that connect two nodes i and j only if the euclidean distance between them is between 4 Angstrom (threshold for only non covalent interactions) and 8 Angstrom (threshold for only significant interactions). The distance between two aminoacids i and j is approssimated by the distance between the Alpha Carbon of the amino acids. The user can modify the only covalent (min) and the only significant (max) threshold distance for PCN construction.
PCN global descriptors (like graph diameter) or local descriptors (like node centrality measures) are useful to model and analyse protein functions. PCN Miner allow the user to identify modules (also called communities or clusters) in protein molecules using three different approaches:
- spectral clustering: extract clusters from a graph with a clustering approach based on the Laplacian matrix eigenvectors following the guidelines given in the paper: A tutorial on spectral clustering [1];
- embedding+clustering: uses one of the embedding algorithm in the GEM library [2] and then apply clustering;
- community detection: uses one of the community detection algorithm in the cdlib library [3].
The main objective of PCN Miner are:
(i) identify the putative allosteric paths and regions in protein structures (that can help the design of allosteric drugs);
(ii) allow the user to hypotize the functional effect of mutations (example variants of SARS CoV-2 Spike Protein);
(iii) recognise funtional domains (communities or clusters) in proteins.
PCN-Miner Supported Algorithms:
- Spectral Clustering: Both Hard (K-Means) and Soft (Fuzzy C-Means) clustering approach used on the eigenvectors of the Laplacian matrix (both normalized or unnormalized form). Is also supported the Shi Malik spectral clustering approach that resolves the generalized eigenvalues problem;
- Embedding + Clustering: Node2vec, HOPE, Laplacianeigenmaps embedding followed by a supported spectral clustering algorithm;
- Community Detection: Louvain, Leiden, Walktrap, Infomap, Asyn FluidC, Greedy Modularity, Spinglass;
- Centrality Measures: Closeness Centrality, Betweenness Centrality, Eigenvector Centrality, Degree Centrality.
Outputs (node centrality, clusters or communities) of the supported algorithms are then plotted on the 3D structure of the protein using PyMol scripts.
The easiest way to install this library is using the setup files on github:
-S.O. Windows:
git clone https://github.com/hguzzi/ProteinContactNetworks.git
cd ProteinContactNetworks
setupWindows.bat
-S.O. Linux-MACOSX:
git clone https://github.com/hguzzi/ProteinContactNetworks.git
cd ProteinContactNetworks
source setupLinux-MacOSX.sh
Note: You may need to convert line endings format of the file setupLinux-MacOSX.sh. You can do it with Notepad++: Open the .sh file and then Edit -> EOL Conversion -> Unix or Macintosh.
You can also install this library with pip:
IMPORTANT: This project depends on PyMOL and GraphEmbeddingMethods, two libraries not available on PyPI.
One easy way to install pymol is using anaconda.
Open the anaconda prompt and type the following command:
conda create -n PCN python=3.8.3
conda activate PCN
conda install -c conda-forge -c schrodinger pymol-bundle
Then we can install GEM library using pip+git:
pip install git+https://github.com/palash1992/GEM.git
Finally we can install this library using TESTPYPI:
pip install --extra-index-url https://pypi.org/simple -i https://test.pypi.org/simple/ pcn
Or with pip:
pip install pcn
Or with pip+git:
pip install git+https://github.com/hguzzi/ProteinContactNetworks.git#egg=pcn
If the software is installed with pip:
conda activate PCN
python
from pcn.tools import pcn_main
pcn_main.main()
If the software is installed with setup files on git:
conda activate PCN
cd pcn
cd tools
python pcn_main.py
If the software is installed with pip:
conda activate PCN
python
from pcn.tools import pcn_gui_main
pcn_gui_main.main()
If the software is installed with setup files on git:
conda activate PCN
cd pcn
cd tools
python pcn_gui_main.py
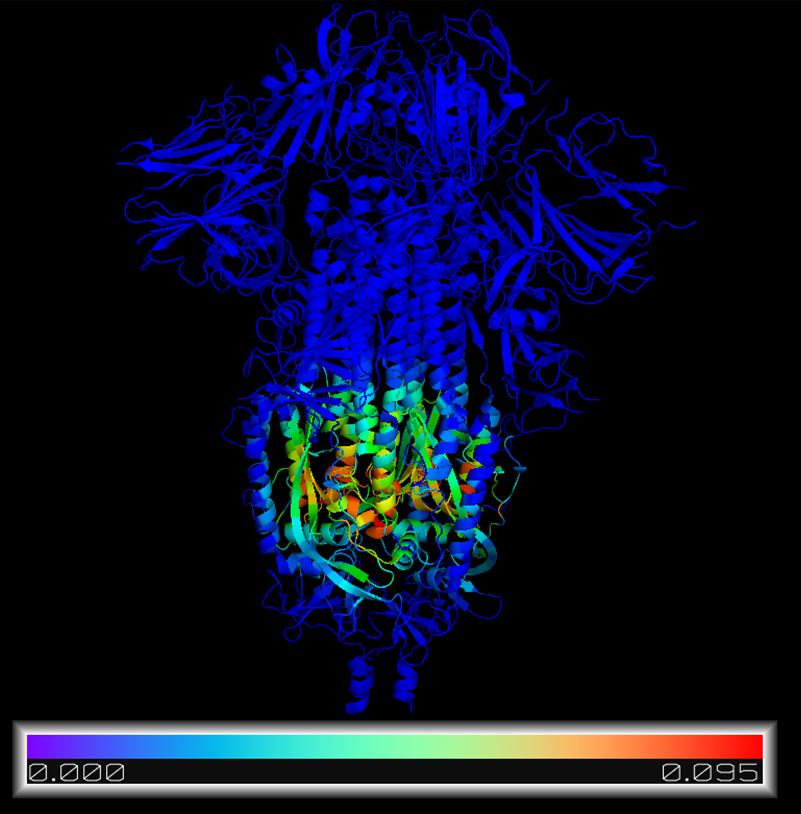
Entry PDB code: 6VXX
Description: SARS CoV 2 Spike protein closed form
Method: Community Detection
Algorithm: Leiden
Number of communities extracted: 20
Method: Centrality Analysis
Algorithm: Eigenvector Centrality
[1] von Luxburg, U. A tutorial on spectral clustering. Stat Comput 17, 395–416 (2007). https://doi.org/10.1007/s11222-007-9033-z;
[2] https://github.com/palash1992/GEM;
[3] G. Rossetti, L. Milli, R. Cazabet. CDlib: a Python Library to Extract, Compare and Evaluate Communities from Complex Networks. Applied Network Science Journal. 2019. DOI:10.1007/s41109-019-0165-9 https://github.com/GiulioRossetti/cdlib