AMatReader is a general adjacency matrix importer plugin for Cytoscape.
Download the app on the Cytoscape app store
Sample adjacency matrix formats can be found in the samples directory.
Below is a sample format for a matrix file:
#Simple tab-delimited adjacency matrix
Node1 Node2 Node3 Node4
Node1 1 4
Node2 1 3 3
Node3 4 3 1
Node4 3 1
More sample adjacency matrix files are available in the samples directory in the parent folder of this repo
Note that lines starting with # are considered comments and are ignored
NOTE: Files imported in either manner MUST use the .adj or .mat suffix to be recognizes as matrix files. To import other filetypes (.csv, .txt, etc.) use the Apps Menu method below
Go to File>Import>Network>File... and select the individual file for import.
OR
Drag multiple .mat/.adj files into the network panel.
Go to Apps> AMatReader> Import Matrix Files. Select the file(s) you want import and hit Import
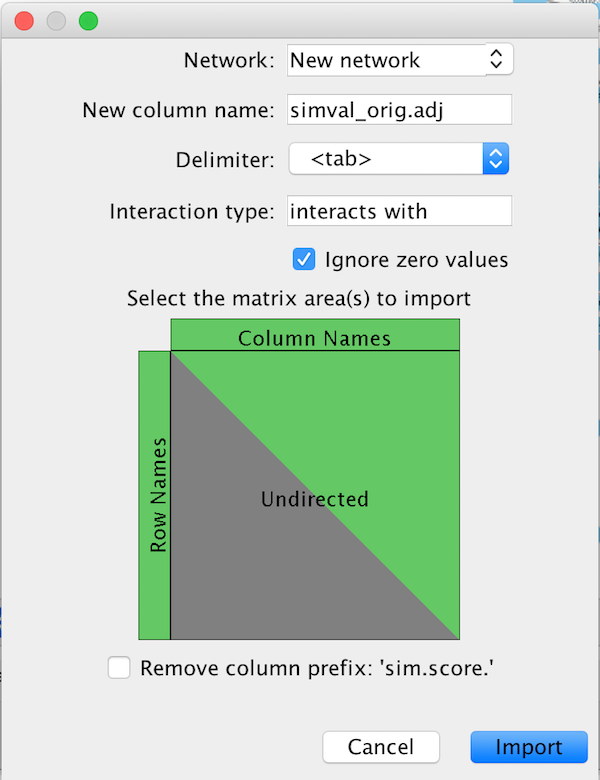
AMatReader will peek at each file and predict the parameters for import.
The settings in this dialog (also available through CyREST) are described below.
| Name | Description |
|---|---|
| files | list of files to be imported |
| delimiter | separator used between values in the matrix (tab, comma, pipe |
| ignoreZeros | whether or not to create edges for zero values in the matrix |
| interactionType | value given to the interaction column in the Cytoscape Edge table |
| rowNames | True if the first column of the matrix specifies node names, False otherwise |
| columnNames | True if the first row of the matrix specifies nodes names, False otherwise |
| undirected | directed-ness of the resulting graph. If True, only the top triangle of the matrix is used |
| removeColumnPrefix | Useful for Matlab/R imports, where column names are prefixed by some description. |
The New column name field in the dialog defines the edge column to be created. In CyREST, the file basename is used as the new column name
A python interface was created to allow importing adjacency matrices into Cytoscape in a RESTful manner. This package uses the CyREST endpoints of AMatReader to import files, pandas DataFrames, and numpy arrays that represent adjacency matrices.
To use the pyCyAMatReader package, follow these steps:
pip install pycyamatreaderStart Cytoscape and make sure the AMatReader app is installed. PyCyAMatReader makes use of the CyREST interface for AMatReader and will only work with Cytoscape 3.6+.
from pyCyAMatReader import AMatReader
_amatreader = AMatReader() # assumes Cytoscape answers at http://localhost:1234data = {
"files": [file1, file2],
"delimiter": "TAB",
"ignoreZeros": True,
"undirected": False,
"interactionName": "interacts with",
"rowNames": True,
"columnNames": True,
"removeColumnPrefix": False
}To import a file as a new Cytoscape network
data['files'] = ['sample.mat']
result = _amatreader.import_matrix(data)Or extend an existing network
data['files'] = ['sample2.mat']
# use the resulting network from the import above
suid = result['suid']
_amatreader.import_matrix(data, suid=suid)You can also now import pandas DataFrames and numpy arrays using aMatReader
NOTE: python dataframes are saved to a temporary file for import. This is not advised for large files. If you have a large file, use the util_numpy helper in py2Cytoscape
To import a numpy matrix that represents an adjacency matrix
mat = np.random.random([5, 5]) # sample 5x5 numpy matrix
# numpy saves matrices without row/column names by default
data = data.update({'columnNames': False,
'rowNames': False,
'undirected': 'true'})
result = _amatreader.import_numpy(mat, data, names=['A', 'B', 'C', 'D', 'E'])To import a pandas DataFrame that represents an adjacency matrix
# pandas does not save with row names by default
data['rowNames'] = False
df = pd.DataFrame(np.random.randint(low=0, high=10, size=(5, 5)),
columns=['A', 'B', 'C', 'D', 'E'])
result = _amatreader.import_pandas(df, data)