In this repository I shortly showcase how Word2Vec can be used for representation learning to create features which represent peptide sequences and secondary structure. Furthermore, the learned representations will be used as input features for a CNN to predict hemolytic activity. The use of Word2Vec to represent biolgical sequences was inspired by [1]. Sequences were taken from [2] and [3]. Since there is a general lack of secondary structure information for proteins, I predicted the secondary structure with [4].
Word2Vec was created with the idea that words in human language derive meaning from the words that are around them. To get numeric features which represent a word, a corpus of sentences is taken and for each word the probability to appear near words in the neighbourhood (neighbourhood probability) is calculated. A neural network is then trained to predict for each word, the neighborhood probability of all other words. This is a proxy-task, because we do not care about the output, but the weights used by the neural network after training. The weights relative for each word are taken as numeric representations or features for these words, will also have the same weights applied to them. The idea behind this is that words that appear near the same word, also have a comparable meaning and thus should have a comparable numeric representation.
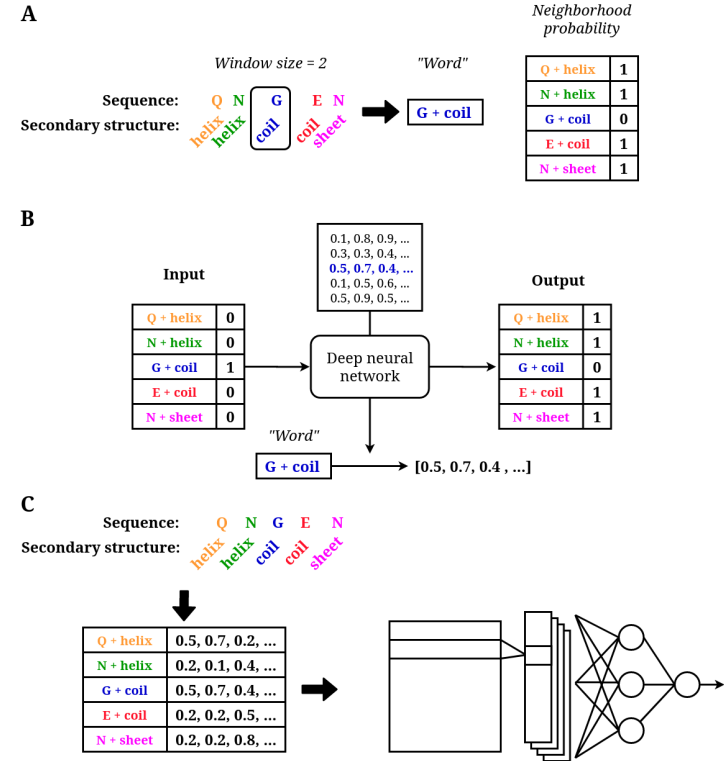
When applied to proteins, the sequence and secondary structure becomes the sentence, with the amino acids and secondary structure as words. To get "words", we split the sequence and structure in n-grams, with in this example only 1-grams. We will use the Word2Vec representations learned for these 1-grams to represent peptides by creating a matrix for each peptide, with per 1-gram in the peptide a row in the matrix with the numeric representation for that 1-gram. To overcome different sequence lengths we zero-pad the matrices.
Figure depicting of how 1-grams are made of a peptide sequence and structure, neighbourhood probability calculations and how a neural network is used to predict this neighborhood probability for each 1-gram. We stop caring about the output and take the weights specific for the input word as representation as input for a CNN./figcaption>- Split sequences and secondary structure of each peptide into 1-grams
- Use Word2Vec to learn numeric representations for each 1-gram
- Visualise numeric representations of 1-gram
- Train and use CNN to predict hemolytic activity of peptides
- R
- CRAN packages in the R notebooks
- Just run the R notebook file in Rstudio
- https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-019-3006-z
- https://academic.oup.com/bioinformatics/article/36/11/3350/5799076
- https://www.nature.com/articles/s41598-020-67701-3#data-availability
- https://github.com/mircare/Porter5
Jeppe Severens jeppeseverens@gmail.com