Our goal is to develop a simple method to qualitatively identify quality libraries for running RNA and DNA-seq. We are using output from the Agilent Tape Stations 2200. Unfortunately, the software provided with the Tape Station provides no means of exporting data, except for picked peaks and gel images. If additional analyses are required (i.e., distribution analysis or tests for normality), then image analysis of exported gels is required. We want to have a set of defined criteria that would allow an unbiased selection of quality libraries.
Here is a brief summary of how to acquire data from the Agilent Tape Station for use with this tool.
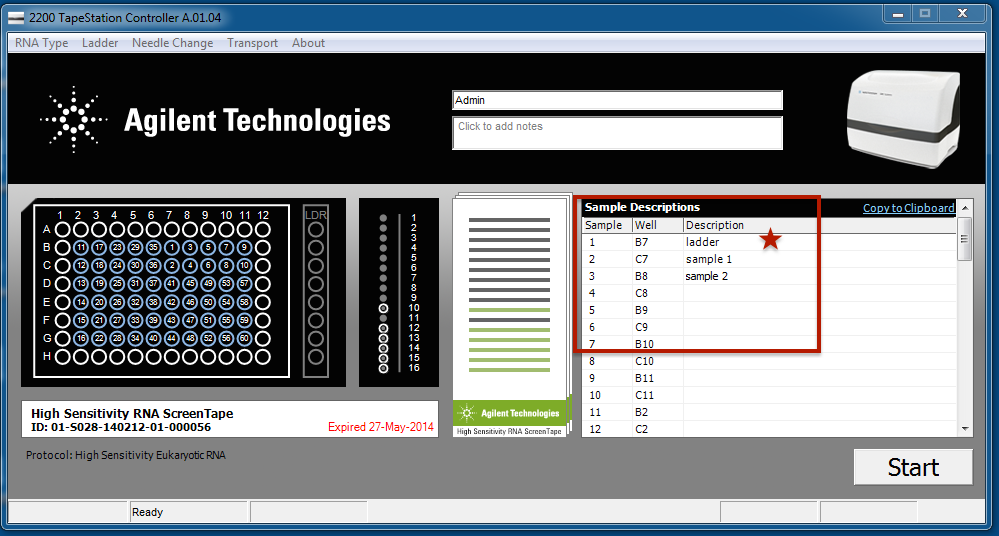
When starting a tape station run, the user has the opportunity to add a description for each sample (red box). These descriptions can be copied and pasted from an Excel worksheet. The tapeAnalyst tool will use this information when generating the report. Currently tapeAnalyst is designed for DNA (cDNA) runs of the Tape Stations, and it is suggested to use a ladder. If you use a ladder make sure to at least add the key work ladder to the description (red star).
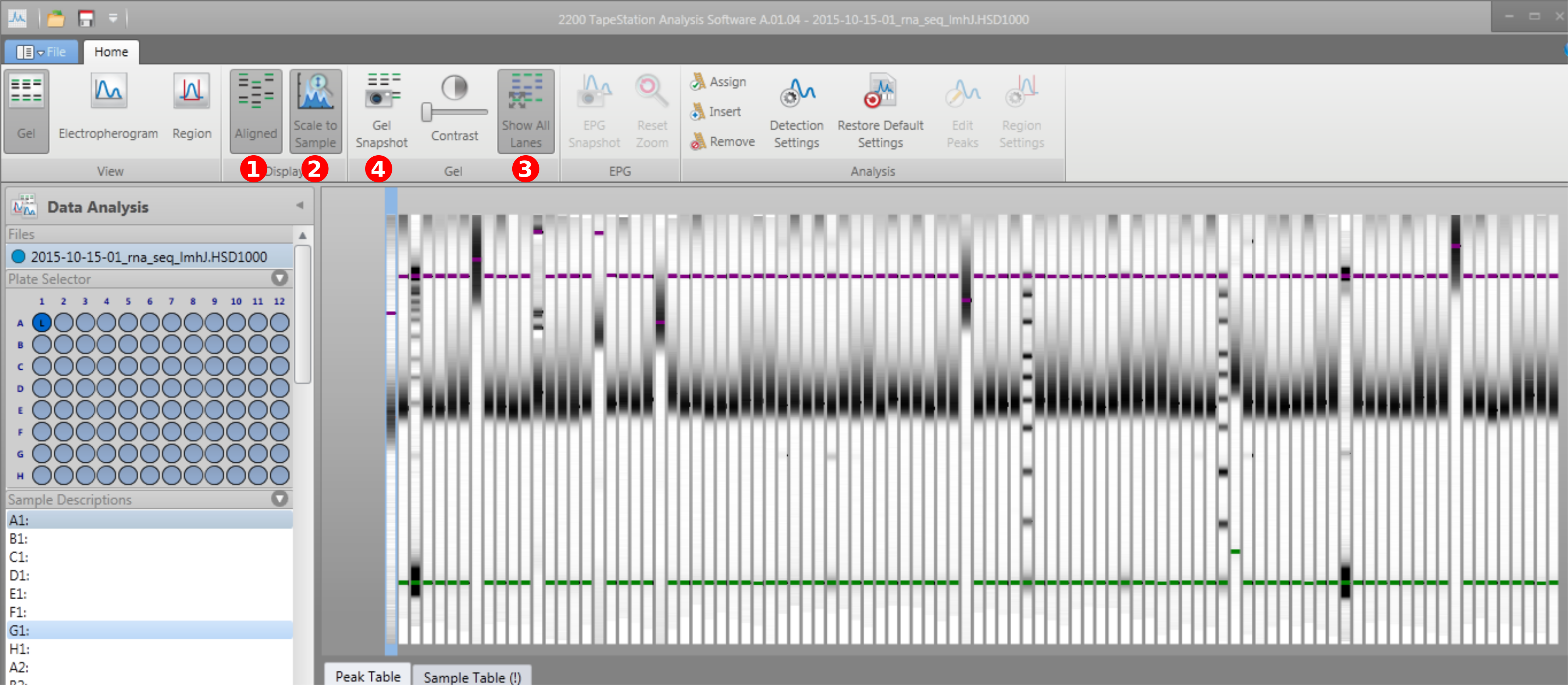
After a successful run the Tape Station, the following steps need to be done to process the data. (1) Align the sample using the dye location. (2) Adjust the scale to each sample. This will make sure the contrast is strong, but will make comparison between samples difficult. If contrast is good, this step can be skipped. (3) Show all lanes at once. (4) Take a gel snap shot.
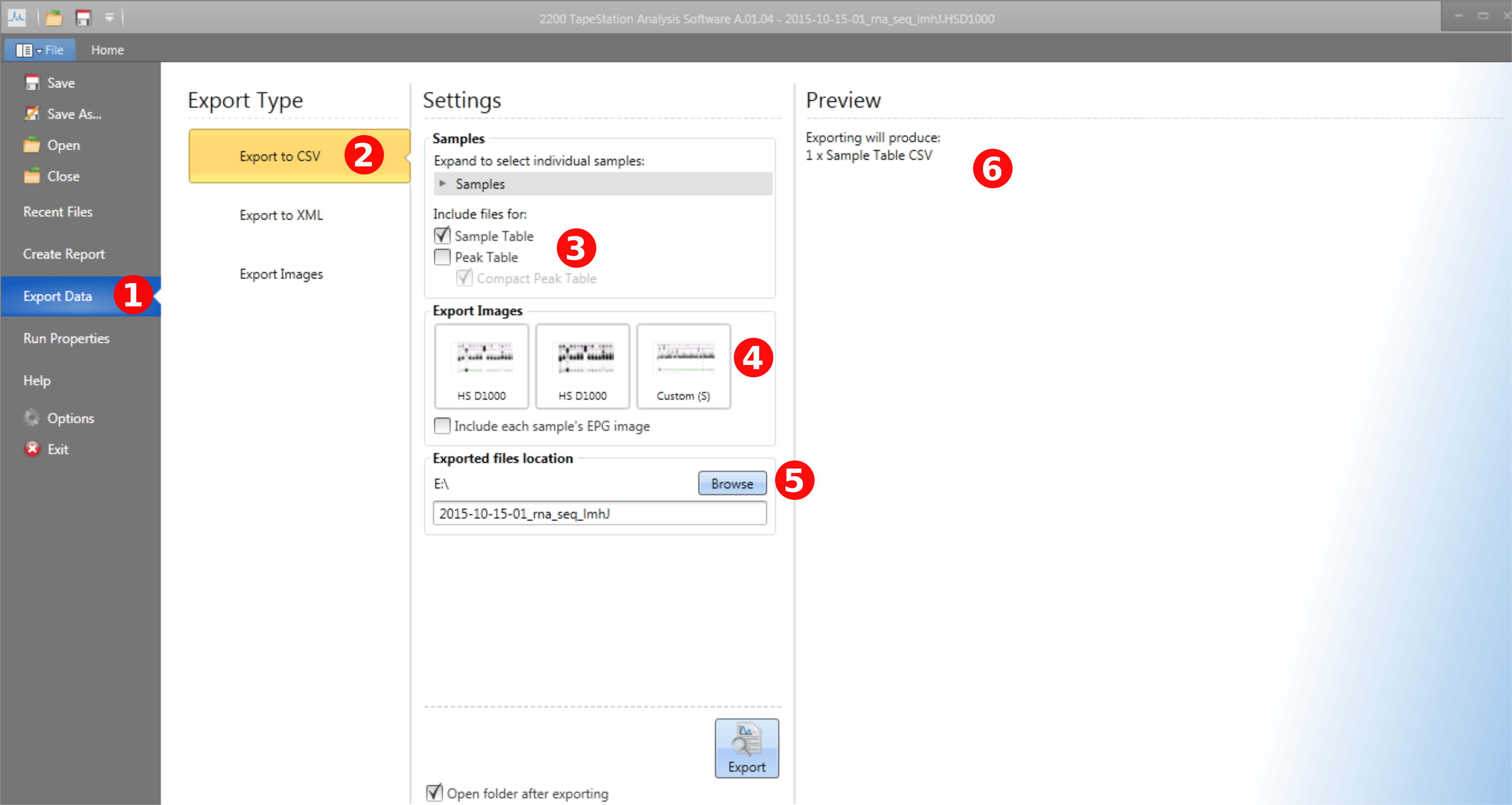
To export data, go to the File tab. (1) Click on the export tab. (2) Select export to CSV. (3) Make sure Sample Table is checked. You can also export the peak table, but this is not used by tapeAnalyst. (4) Make sure to select the gel image of the entire plate. Note sometimes is takes 15-30 seconds for all of the images to show up here. If the full plate image does not show up make sure you did (4) in the processing section. (5) Select the location you want to export. (6) Will list all of the files you are going to export. Here I only have the Sample Table, make sure the gel image is also listed here before export.
Installation script has not been completed. For now you can do the following.
mkdir $HOME/devel
cd $HOME/devel
git clone https://github.com/jfear/tapeAnalyst.gitYou can run this before trying to run tapeAnalyst.
export PYTHONPATH=$PYTHONPATH:$HOME/devel/tapeAnalystOr you can make the change to your .profile or .bashrc to make the change permanent.
echo "export PYTHONPATH=$PYTHONPATH:$HOME/devel/tapeAnalyst" >> ~/.profilecd $HOME/devel/tapeAnalyst
make installtapeAnalyst is a basic command line tool. See command line options for more details by running the tool with -h.
cd $HOME/devel/tapeAnalyst/bin
analyzeTapeStationPng -h