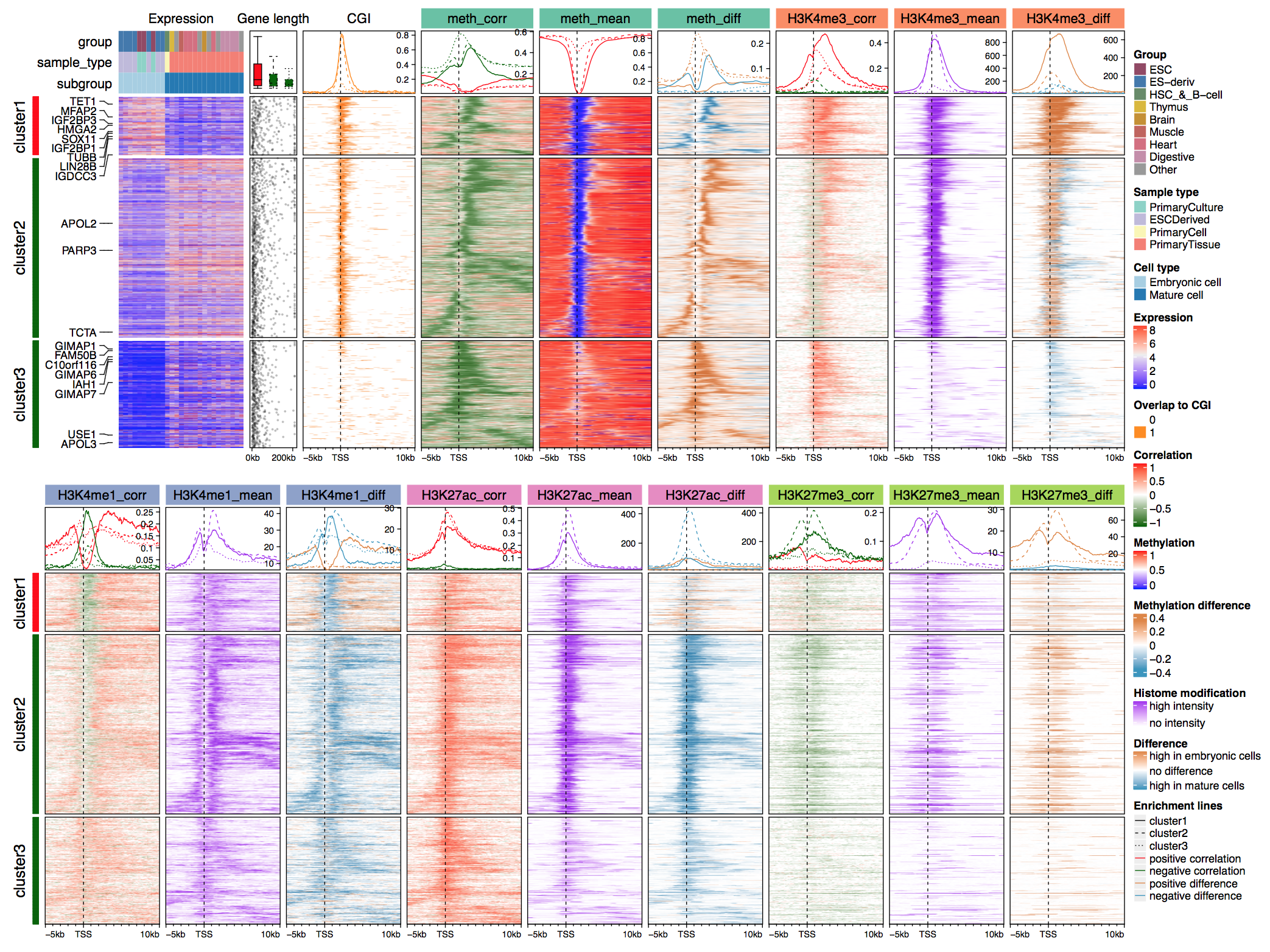
Enriched heatmap is a special type of heatmap which visualizes the enrichment of genomic signals on specific target regions. It is broadly used to visualize e.g. how histone marks are enriched to specific sites.
There are several tools that can make such heatmap (e.g. ngs.plot or deepTools). Here we implement Enriched heatmap by ComplexHeatmap package. Since this type of heatmap is just a normal heatmap but with some fixed settings, with the functionality of ComplexHeatmap, it would be much easier to customize the heatmap as well as concatenating a list of heatmaps to show correspondance between differnet data sources.
Zuguang Gu, et al., EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations, 2018. BMC Genomics. link
EnrichedHeatmap is available on Bioconductor, you can install it by:
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("EnrichedHeatmap")
If you want the latest version, install it directly from GitHub:
library(devtools)
install_github("jokergoo/ComplexHeatmap")
install_github("jokergoo/EnrichedHeatmap")
Like other tools, the task involves two steps:
- Normalize the accosiations between genomic signals and target regions to a matrix.
- Draw heatmaps.
mat1 = normalizeToMatrix(H3K4me3, tss, value_column = "coverage",
extend = 5000, mean_mode = "w0", w = 50)
mat2 = normalizeToMatrix(meth, tss, value_column = "meth", mean_mode = "absolute",
extend = 5000, w = 50, background = NA, smooth = TRUE)
partition = kmeans(mat1, centers = 3)$cluster
lgd = Legend(at = c("cluster1", "cluster2", "cluster3"), title = "Clusters",
type = "lines", legend_gp = gpar(col = 2:4))
ht_list = Heatmap(partition, col = structure(2:4, names = as.character(1:3)), name = "partition",
show_row_names = FALSE, width = unit(3, "mm")) +
EnrichedHeatmap(mat1, col = c("white", "red"), name = "H3K4me3", row_split = partition,
top_annotation = HeatmapAnnotation(lines = anno_enriched(gp = gpar(col = 2:4))),
column_title = "H3K4me3") +
EnrichedHeatmap(mat2, name = "methylation",
top_annotation = HeatmapAnnotation(lines = anno_enriched(gp = gpar(col = 2:4))),
column_title = "Methylation") +
Heatmap(log2(rpkm+1), col = c("white", "orange"), name = "log2(rpkm+1)",
show_row_names = FALSE, width = unit(5, "mm"))
draw(ht_list, main_heatmap = "H3K4me3", gap = unit(c(2, 10, 2), "mm"))
Also when signals are discreate values. E.g. chromatin states:
Actually you can generate rather complex heatmaps:
MIT @ Zuguang Gu