This repository serves as a backup for utility scripts created by José Pereira, on the scope of Protein Conformation Prediction. Most scripts are deployed using a TKinter GUI. It was last updated on 26 April 2018. The following sequence of scripts facilitates the process of:
- Downloading PT trajectory data from Mongo;
- Run clusteriazation;
- Transform the selected coarse-grained structure to 'all'-atoms model;
- Run complete molecular dynamics simulations;
- Analyze the generated data.
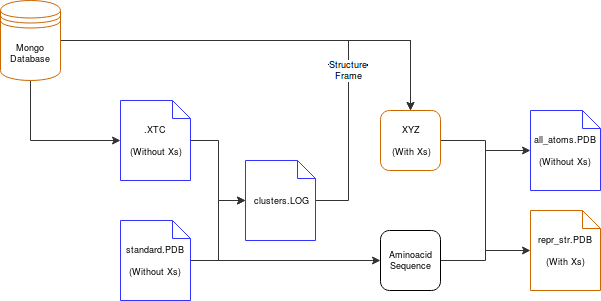
This script automates the process of downloading data from MONGO database as XTC trajectory files. The XTC trajectory frames are then clustered based on RMSD, and, if requested, the plot depicting Energy values vs RMSD is displayed to the user for Cluster extraction selection. The selected clusters' representative structure is then retrieved and transformed into an all-atom model for MD analysis.
The script requires the pre-installment of the following dependencies:
- Tkinter (including tkMessageBox)
- ttkThemes
- MatPlotLib
- MDTraj
- PyMongo
- ProtoSyn (Not publicly avaliable)
- Numpy
- Gui (Avaliable from this repository)
For the full usage of this script, the following files are required:
- PDB structure file of the original protein for RMSD comparison
- A Parallel Tempering Database at a MONGO server
The script outputs the following files to the current working directory ('traj' is the default name):
- traj_all_atoms.pdb > Representative structure of the selected cluster in all_atoms form
- traj.log > Log file depicting all the clusters formed
- clusters.png > Figure of the RMSD vs Energy graph
- traj_clusters.pdb > Trajectory where the PDB frames are the representative structures of each cluster (without 'X's)
- traj_repr_str.pdb > Representative stucture of the selected cluster in caterpillar form (with 'X')
- others > Mostly leftover files, but can be usefull in certain cases: traj.xtc, traj.xvg, rmsd-dist.xvg, traj.xpm
This script automates the process of running and MD simulation. for each simulation step, the user can select the number of steps (or simulation time by multiplying by dt = 0.002), aswell as the frequency of data storage. The MD process is comprised of 7 automated steps:
- Preparation (Adds bounding box and creates topology files);
- Minimization 1 (Without solvent);
- Solvation (Adds spc216 water molecules);
- Minimization 2 (With solvent);
- NVT equilibration;
- NPT equilibration;
- Data collection.
The script requires the pre-installment of the following dependencies:
- Tkinter (including tkMessageBox)
- ttkThemes
- Gui (Avaliable from this repository)
For the full usage of this script, the following files are required:
- An .mdp storage folder (Avaliable from this repository)
- A starting .pdb structure
This script intends to automize the process of analysing GROMACS MD results. Based on user requests, it can perform up 4 tasks:
- Plot RMSD vs Frame 0 of the MD trajectory
- Plot RMSD vs Original protein
- Plot Energy potential as a function of simulation time
- Plot Gyration degrees (total and in discrete axis) as a function of simulation time
The script requires the pre-installment of the following dependencies:
- Tkinter (including tkMessageBox)
- ttkThemes
- MatPlotLib
- Gui (Avaliable from this repository)
For the full usage of the script, the following GROMACS files are required:
- TPR topology
- XTC trajectory
- EDR energy file
- PDB structure file of the original protein for RMSD comparison