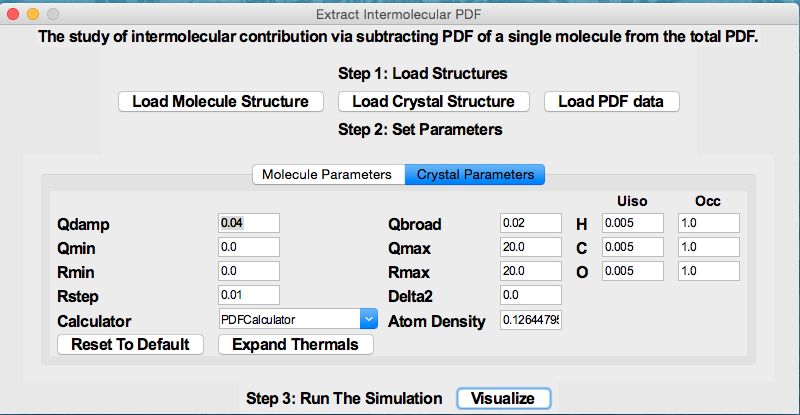
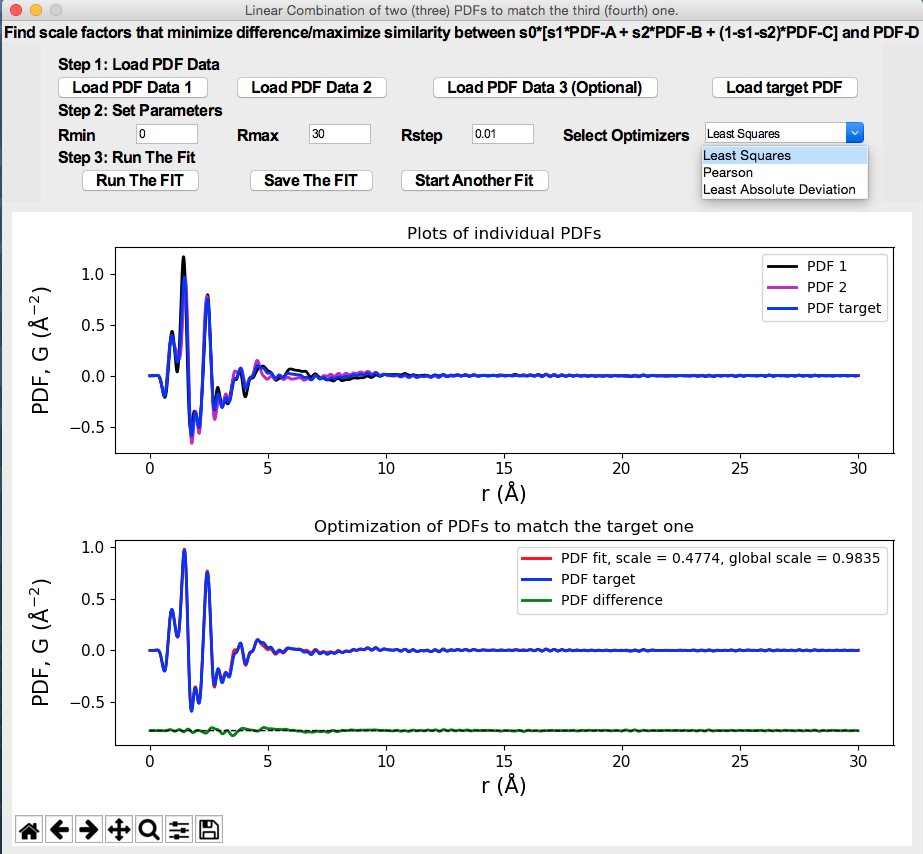
(A video demo about installation and usage is available at https://www.youtube.com/watch?v=lAFZ5VYEH1g).
Python GUI program for analyzing organic pair distribution function (PDF) data collected at synchrotron and/or laboratory X-ray sources. It uses DiffPy-CMI (http://www.diffpy.org/products/diffpycmi/index.html) as a backend for simulation of PDFs. Currently it supports (1) The study of intermolecular interaction (e.g. hydrogen bonds) by subtracting out the scattering signal of single molecule(s) in real space. (2) The PDF model fit of the crystalline organic compound using the method proposed by Prill et al. (J. Appl. Cryst., 48, 171-178, 2015.) (3) The phase quantification of physical mixtures of organics. (4) Generate Score/Scree plots based on Principle Component Analysis.
If you like the program and use it in your own work, it would be appreciated if you cite the following papers.
(1) Chenyang Shi, “xINTERPDF: a GUI program for analyzing intermolecular pair distribution functions of organic compounds from X-ray total scattering data”, under review, J. Appl. Cryst., 2018.
(2) Pavol Juhás, Christopher L. Farrow, Xiaohao Yang, Kevin R. Knox, and Simon J. L. Billinge, “Complex modeling: a strategy and software program for combining multiple information sources to solve ill posed structure and nanostructure inverse problems”, Acta Crystallogr. A, 71, 562-568, 2015.
A quick view of the program is shown below.
(See also Slides 11-14 in User Guide available at https://github.com/curieshicy/xINTERPDF/blob/master/xINTERPDF_User_Guide_20180615.pdf)
xINTERPDF can be installed on Linux and macOS computers. The easiest way to install it is through conda. Here is an example of installing it on macOS 10.10.3.
(1) Download Anaconda Distribution for macOS at https://www.anaconda.com/download/?lang=en-us#macos. Select Python 2.7 version to install.
(2) Invoke a terminal, type conda config --get channels to check any channels that have been added. diffpy is required. If you don’t see it, type conda config --add channels diffpy to add it.
(3) Type conda create –c curieshicy –n xinterpdf xinterpdf to install the xINTERPDF program.
(4) Once the installation is complete. Type source activate xinterpdf to start the virtual environment and xinterpdf to invoke the main window of xINTERPDF
(Alternatively) If conda install fails, one may download the raw files (Logo.gif and cli.py) at https://github.com/curieshicy/xINTERPDF/tree/master/Conda_Recipe_macOS_Linux/Conda_Build_Recipe_macOS/xinterpdf (macOS) or https://github.com/curieshicy/xINTERPDF/tree/master/Conda_Recipe_macOS_Linux/Conda_Build_Recipe_Linux/xinterpdf (Linux). To start the program, in a terminal, navigate to the folder where you put both files, and type python cli.py to invoke the main window. Make sure you have installed Diffpy-CMI, matplotlib (2.0.2) and Scikit-Learn (0.19.1). Follow http://www.diffpy.org/products/diffpycmi/index.html to install DiffPy-CMI. If you have conda, matplotlib can be installed by conda install matplotlib=2.0.2; Scikit-Learn can be installed by conda install scikit-learn=0.19.1
A user guide for xINTERPDF can be found at https://github.com/curieshicy/xINTERPDF/blob/master/xINTERPDF_User_Guide_20180615.pdf. It contains an overview of the program, a step-by-step installation guide, an explaination of techincal terms and a demonstration on three examples. The example files are available at https://github.com/curieshicy/xINTERPDF/tree/master/Examples.
Questions and suggestions are welcome. You may contact me at cs3000@columbia.edu. Thanks for considering using xINTERPDF.