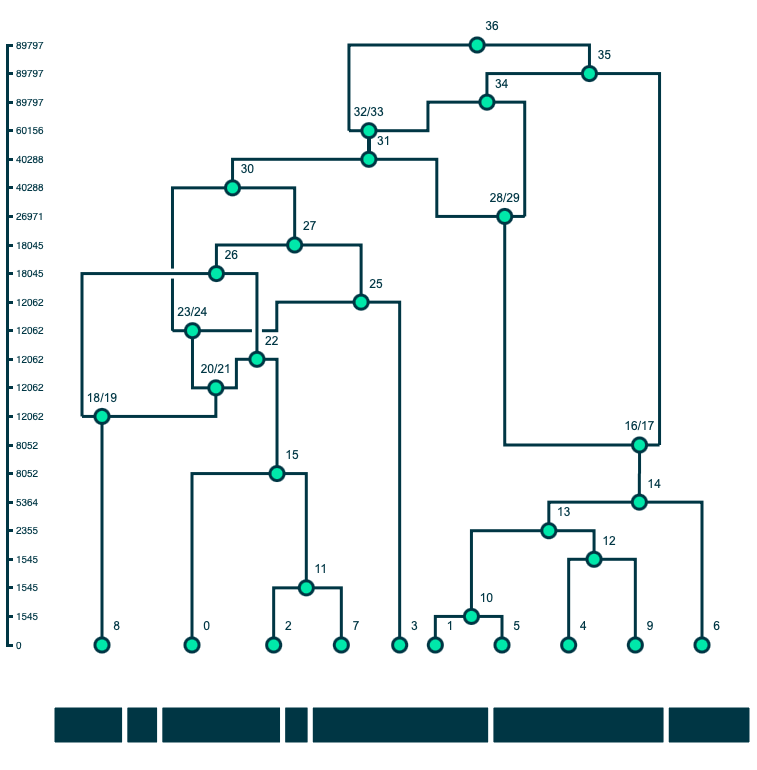
A method for drawing ancestral recombination graphs from tskit tree sequences in Python using D3.js. ARGs are plotted using a D3's force layout. All nodes have a fixed position on the y-axis set by fy. Sample nodes have a fixed position on the x-axis set by fx; the ordering of the sample nodes comes from the first tree in the tskit tree sequence (this is not always the optimal ordering but is generally a good starting point for plotting). The x positions of other nodes are set by a force simulation where all nodes repel each other countered by a linkage force between connected nodes in the graph.
Users can click and drag the nodes (including the sample) along the x-axis to further clean up the layout of the graph. The simulation does not take into account line crosses, which can often be improved with some fiddling. Once a node has been moved by a user, its position is fixed with regards to the force simulation.
pip install tskit_arg_visualizer
This package loads D3.js using a CDN, so requires access to the internet.
import msprime
import random
import tskit_arg_visualizer
# Generate a random tree sequence with record_full_arg=True so that you get marked recombination nodes
ts_rs = random.randint(0,10000)
ts = msprime.sim_ancestry(
samples=3,
recombination_rate=1e-8,
sequence_length=3_000,
population_size=10_000,
record_full_arg=True,
random_seed=ts_rs
)
d3arg = tskit_arg_visualizer.D3ARG.from_ts(ts=ts)
d3arg.draw(
width=750,
height=750,
edge_type="ortho"
)
The above code can be run in three ways: terminal, Jupyter Notebook, or JupyterLab. For Jupyter Notebook and JupyterLab, you will need to add the following code block to the top of the document to properly load D3.js.
%%javascript
require.config({
paths: {
d3: 'https://d3js.org/d3.v7.min'
}});
require(["d3"], function(d3) {
window.d3 = d3;
});
%%javascript
var script = document.createElement('script');
script.type = 'text/javascript';
script.src = 'https://d3js.org/d3.v7.min.js';
document.head.appendChild(script);
The draw() function for a D3ARG object has multiple parameters that give users a small amount of customization with their figures:
def draw(
self,
width=500,
height=500,
tree_highlighting=True,
y_axis_labels=True,
y_axis_scale="rank",
edge_type="line",
variable_edge_width=False,
include_underlink=True,
sample_order=[]
):
"""Draws the D3ARG using D3.js by sending a custom JSON object to visualizer.js
Parameters
----------
width : int
Width of the force layout graph plot in pixels (default=500)
height : int
Height of the force layout graph plot in pixels (default=500)
tree_highlighting : bool
Include the interactive chromosome at the bottom of the figure to
to let users highlight trees in the ARG (default=True)
y_axis_labels : bool
Includes labelled y-axis on the left of the figure (default=True)
y_axis_scale : string
Scale used for the positioning nodes along the y-axis. Options:
"rank" (default) - equal vertical spacing between nodes
"time" - vertical spacing is proportional to the time
"log_time" - proportional to the log of time
edge_type : string
Pathing type for edges between nodes. Options:
"line" (default) - simple straight lines between the nodes
"ortho" - custom pathing (see pathing.md for more details, should only be used with full ARGs)
variable_edge_width : bool
Scales the stroke width of edges in the visualization will be proportional to the fraction of
sequence in which that edge is found. (default=False)
include_underlink : bool
Includes an "underlink" for each edge gives a gap during edge crosses. This is currently only
implemented for `edge_type="ortho"`. (default=True)
sample_order : list
Sample nodes IDs in desired order. Must only include sample nodes IDs, but does not
need to include all sample nodes IDs. (default=[], order is set by first tree in tree sequence)
"""
A quick note about line_type="ortho" (more details can be found within pathing.md) - this parameter identifies node types based on msprime flags and applies pathing rules following those types. Because of this, "ortho" should only be used for full ARGs with proper msprime flags and where nodes have a maximum of two parents or children. Other tree sequences, including simplified tree sequences (those without marked recombination nodes marked) should use the "line" edge_type.
Often ARGs from real data can be quite large and unwieldy to visualize all at once. In these instances, we may want to look at the local connections around a specific node of interest.
def draw_node(
self,
node,
width=500,
height=500,
degree=1,
y_axis_labels=True
):
"""Draws a subgraph of the D3ARG using D3.js by sending a custom JSON object to visualizer.js
Parameters
----------
node : int
Node ID that will be central to the subgraph
width : int
Width of the force layout graph plot in pixels (default=500)
height : int
Height of the force layout graph plot in pixels (default=500)
degree : int
Number of degrees above and below the central node to include in the subgraph (default=1)
y_axis_labels : bool
Includes labelled y-axis on the left of the figure (default=True)
y_axis_scale : string
Scale used for the positioning nodes along the y-axis. Options:
"rank" (default) - equal vertical spacing between nodes
"time" - vertical spacing is proportional to the time
"log_time" - proportional to the log of time
"""
This function is very similar to the standard draw(), but you need to provide a node ID which will be the center of the subgraph. At the moment, it also doesn't have quite as many optional styling parameters; graphs are always drawn with tree_highlighting=False, y_axis_scale="rank" and edge_type="line", variable_edge_width=False, and include_underlink=True.
The leftmost button in the visualizer's dashboard provides options for downloading the figure in three formats: JSON, SVG, or PNG. Each figure is actually just a JSON object that D3.js interprets and plots to the screen (see plotting.md for more information about this object). All files are saved as "tskit_arg_visualizer.x", where x is the respective file format.
The JSON file includes all of the information needed to replicate the figure in a subsequent simulation using the following code blocks:
import json
import tskit_arg_visualizer
arg_json = json.load(open("tskit_arg_visualizer.json", "r"))
tskit_arg_visualizer.draw_D3(arg_json=arg_json)
Alternatively, you can pass the JSON into a D3ARG object constructor, which then gives you access to all of the D3ARG object methods, such as drawing and modifying node labels.
d3arg = tskit_arg_visualizer.D3ARG.from_json(json=arg_json)
d3arg.draw()
Lastly, the PNG and SVG files are static files and directly match the current view of the visualizer but without interactivity. A small note, opening the SVG in Adobe Illustrator does not properly import all styles (only inline styles). Though all styles can be manually added or changed within Illustrator, this can be tedious. Styles are properly load when opening in a web browser.
d3arg.set_node_labels(labels={0:"James",6:""})
Node labels can be changed using the set_node_labels() function. The example above will change the labels of Nodes 0 and 6 to "James" and "", respectively. An empty string is equivalent to removing the label of that specific node. Labels are always converted to strings. You can then redraw d3arg with the updated labels. If you wish to go back to the default labelling scheme, you run:
d3arg.reset_node_labels()
The energy of the force layout simulation reduces overtime, causing the nodes to lose speed and settle into positions. Additionally, anytime the user moves a node by dragging, its new position becomes fixed and there on out unchanged by the simulation. The "Reheat Simulation" button in the top left of each figure unfixes the positions of all nodes except for the sample nodes at the tips, and gives a burst of energy to the simulation to allow the nodes to find new optimal positions. This feature is most useful when the starting sample node positions are not optimal; the user can rearrange them and then reheat the simulation to see if that helps with untangling.