New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Inverse Transform? #44
Comments
|
I have given the issue some thought, and have at least a theoretical back of the envelope sketch of how to do it. I don't have any immediate plans in code right now since there are a number of other items that currently have priority. If you would be interested in implementing it yourself please email me and I can try to outline what would be involved. |
|
Hi! |
|
Thanks @danielnilssonjj . Email me at leland.mcinnes@gmail.com and I can try to sketch the process for you. |
|
Wondering if there has been any progress on this @danielnilssonjj @lmcinnes ? |
|
Yes, if you check the 0.4dev branch you'll find an implementation. It isn't the fastest, but it should work. |
|
Thanks for highlighting this. It is good to see people exercising this code. I'll have to dig in and find out what is going on here. I can't promise a quick resolution, but it is definitely important to get this working. |
Hi, I was wondering if the "inverse_transform" function is disabled. |
|
You need the latest version, which is currently on in pre-release. That means it is not the default on PyPI, but can be accessed using the --pre flag. Thus you can do to install that version. |
|
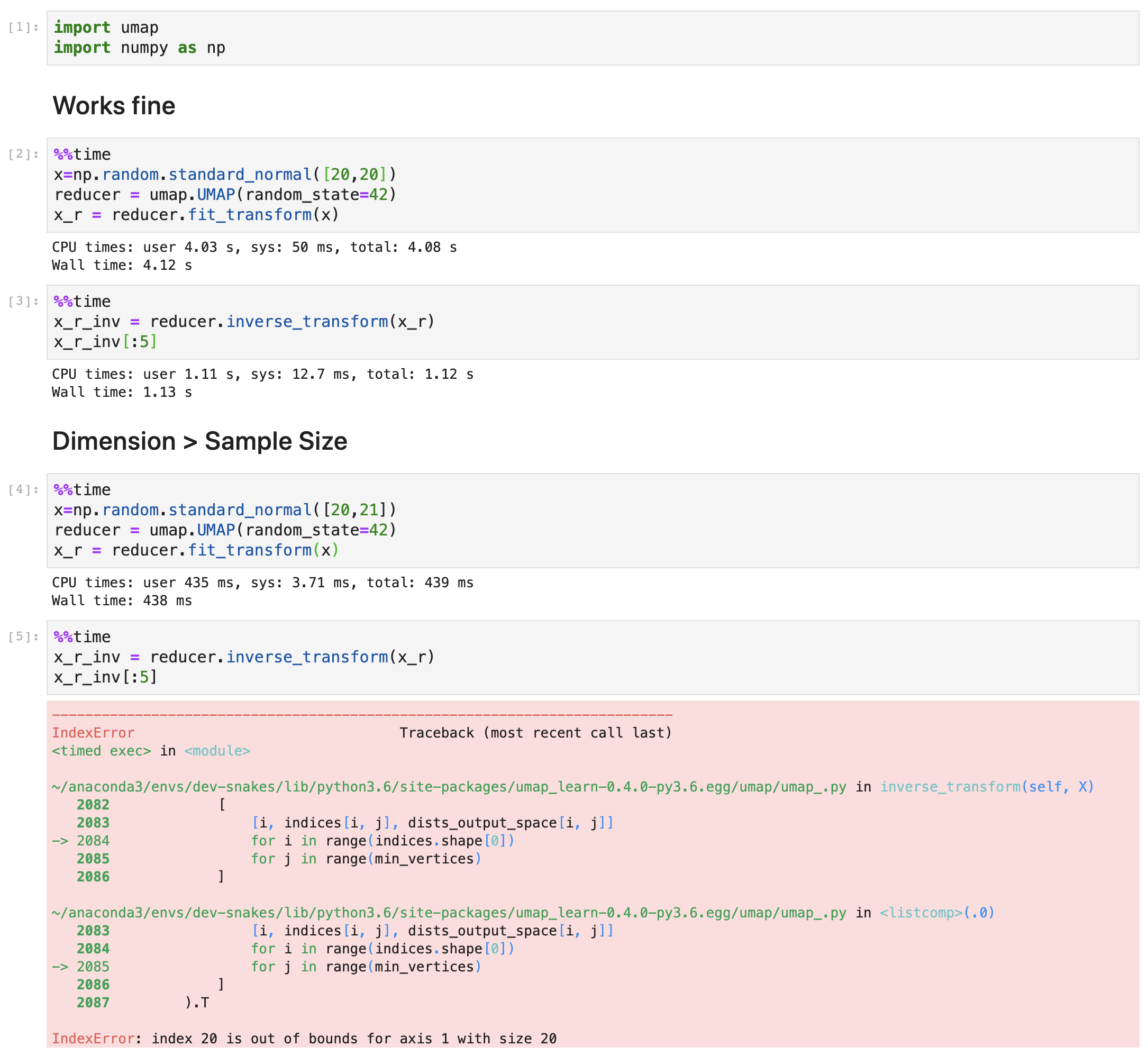
Hi, I was wondering if the problem stated by @ahsanMah is solved. I'm trying to apply the "inverse_transform" but I'm encountering the same problem: The data I'm working with has 53 samples of 7168 features (53,7168). Thanks to UMAP, I achieve I'm using the version "umap-learn==0.4.0rc1". Thanks in advance, I really appreciate all the work done! |
|
The current master branch has the problem resolved, but no release to PyPI has the fix yet. You can clone from github and install from that and it should work. |
Hi, thanks for the quick response! I have downloaded the current master branch and installed UMAP manually as the readme suggests (installing the dependencies and then using setup.py), but the error keeps appearing. Is there something that I'm missing? Thanks in advance! |
|
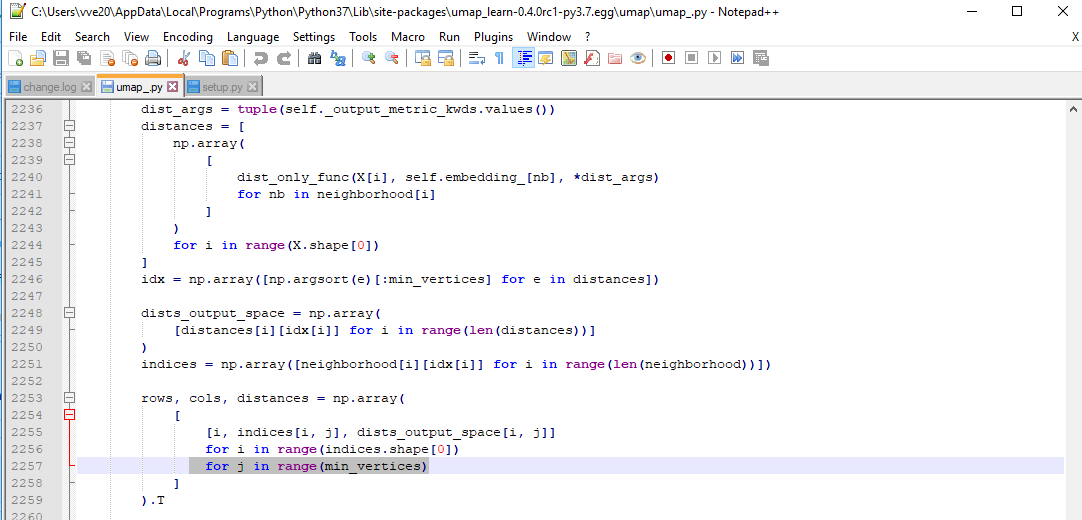
Okay, I'll try to take a look and see if I can figure out the right fix here. |
|
I believe I have a potential fix. I haven't built a reproducer yet, so I can't test right now. If you would like to try with the current master and see if this resolves the issue I would appreciate it. |
Hi, trying the current master branch and installing it manually seems to work. I have achieved to pass from data with shape (53,7168) to 2D space and then use 3 new samples in the 2D space back into the input space with the output shape of (3,7168). Thanks a lot for looking into it, I appreciate it! Keep the amazing work! :) |
|
Thanks -- hopefully others can also verify that it is working for them now.
…On Thu, Mar 12, 2020 at 11:50 AM paudom ***@***.***> wrote:
I believe I have a potential fix. I haven't built a reproducer yet, so I
can't test right now. If you would like to try with the current master and
see if this resolves the issue I would appreciate it.
Hi, trying the current master branch and installing it manually seems to
work.
[image: Captura de pantalla 2020-03-12 a las 16 45 44]
<https://user-images.githubusercontent.com/37597137/76539450-168b2d00-6481-11ea-9bb5-27105fd9f217.png>
I have achieved to pass from data with shape (53,7168) to 2D space and
then use 3 new samples in the 2D space back into the input space with the
output shape of (3,7168).
Thanks a lot for looking into it, I appreciate it! Keep the amazing work!
:)
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#44 (comment)>, or
unsubscribe
<https://github.com/notifications/unsubscribe-auth/AC3IUBL2HFSKCQ3O7A252CLRHEADFANCNFSM4ERGGMRA>
.
|
|
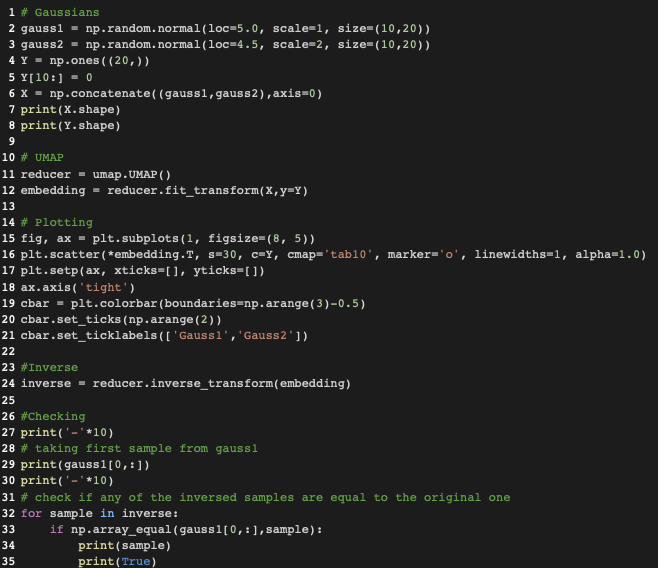
Hi @lmcinnes, I would like to know: If a data point is projected using UMAP into 2D space and then use the inverse_transform on that same point, should it return the same original datapoint? Or I'm interpreting wrong how the inverse transform should work? I'm trying to recover datapoints once UMAP has been applied and I haven't been able to do it. |
|
The inverse transform, like the transform, is stochastic in nature. That means that in practice you can't be guaranteed to get the same point back. Ideally you will get a point very close to it. |
|
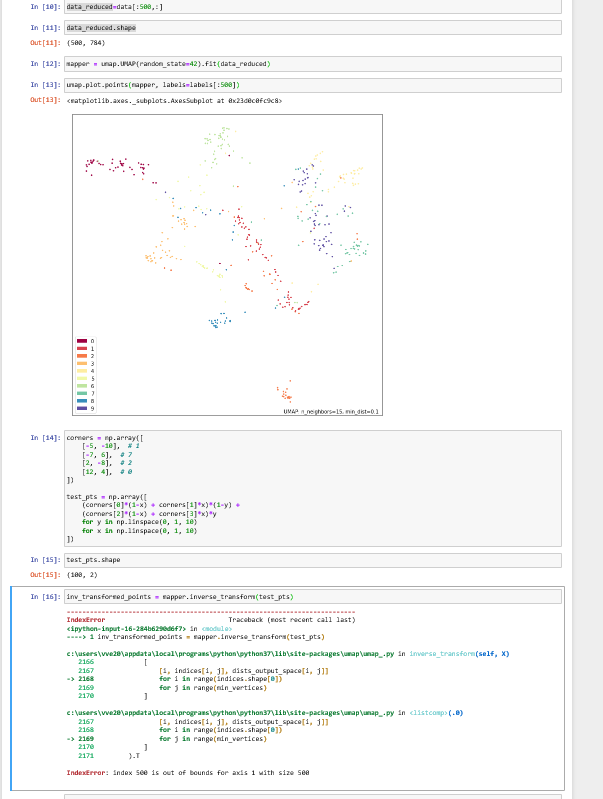
I continued along the idea of @paudom to use the inverse UMAP after UMAP in order to measure the reconstruction error, and to see to what "%var explained" this reconstruction error would correspond to if it were a PCA (https://stats.stackexchange.com/questions/184603/in-pca-what-is-the-connection-between-explained-variance-and-squared-error). This, to give an estimate of the extent to which one grasps "the whole picture" when looking at a 2D or 3D UMAP projection. For example, below 80% "var explained" one would need to be cautious when relying on a UMAP result. As you indicate @lmcinnes, this reconstruction error embeds a margin due to the stochastic nature of the inverse UMAP. My question is: is there then a way to appreciate how to rectify the reconstruction error ? as a mean to appreciate how representative of the data a UMAP result is PS: to partially answer, I investigated the behavior of the UMAP-inverseUMAP reconstruction error, starting with columns of uniform random numbers (a case of limited dimension reduction as there is no major pattern). It shed some light :
|


Hi!
Cool project! If there any plan to implement an inverse op: embedding -> data ? Something like
http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html#sklearn.decomposition.PCA.inverse_transform
Thank you!
The text was updated successfully, but these errors were encountered: