We made all the tools and data used in the research publicly available in order to claim "Available" and "Reusable" batches. This artifact consists of the open-source versions of the tool (PyEvolve) and two kinds of data sets that are used to evaluate the tool. We have readily installed the tool on a VirtualBox VM image and included the evaluation subjects for the reviewer’s convenience. The git repositories of the tools contain detailed instructions for building and using the tools, allowing the public to reuse them.
Note 1: The VirtualBox VM image is a 19GB file. The time it takes to download this file is determined by the internet connection speed. Therefore, we would like to kindly request that reviewers begin downloading this file prior to beginning the review process (check Step 1.2 of Section-1). Furthermore, reviewers must have at least 40 GB of free space on their computer in order to load the file into VirtualBox.
Note 2: We have generated a VirtualBox image on a Mac computer with an Intel chip. The evaluation process heavily relies on the ability to load and run the virtual machine, which may pose challenges as VirtualBox is known to have specific compatibility issues with certain operating systems. To mitigate this, we recommend the reviewer have Mac computer with an Intel chip to run the attached image.
Note 3: We tested the compatibility of the VM image and evaluation instructions on a different machine (Mac-Intel chip), not the one used for creating the image, in order to ensure that all steps will run without interruption.
In Section 1, we first presented the steps to execute the tool in VirtualBox VM image which can be done in under 30 minutes. In section 2, we described our public datasets. The public may access all of these resources (tool and data) through our primary website or archived repositories at Zenodo.
We release one tool
- PyEvolve - a tool that automatically transplants code change patterns to Python software systems
We made two types of large datasets available.
- More than 40,000 code transformation trials were used to evaluate PyEvolve. This was achieved using a dataset of change patterns collected from real repositories. The dataset is publically available.
- PyEvolve atomatically transplanted code change patterns by submitting 40 pull requests to open source repositories. We made the list of the pull requests public.
1. Tool - PyEvolve
Step 1.1: If you do not already have VirtualBox installed, please use this link (https://www.virtualbox.org) and follow the instructions in the link to install VirtualBox on your computer.
Step 1.2: We offer a VM image with all the tools and data pre-installed. The image (named pyevolve.ova) can be downloaded from the links provided below. The image file is 19GB in size and may take some time to download depending on your internet connection speed. We have provided a few links below in case the first one does not work. You can get the image by clicking on one of the links provided below.
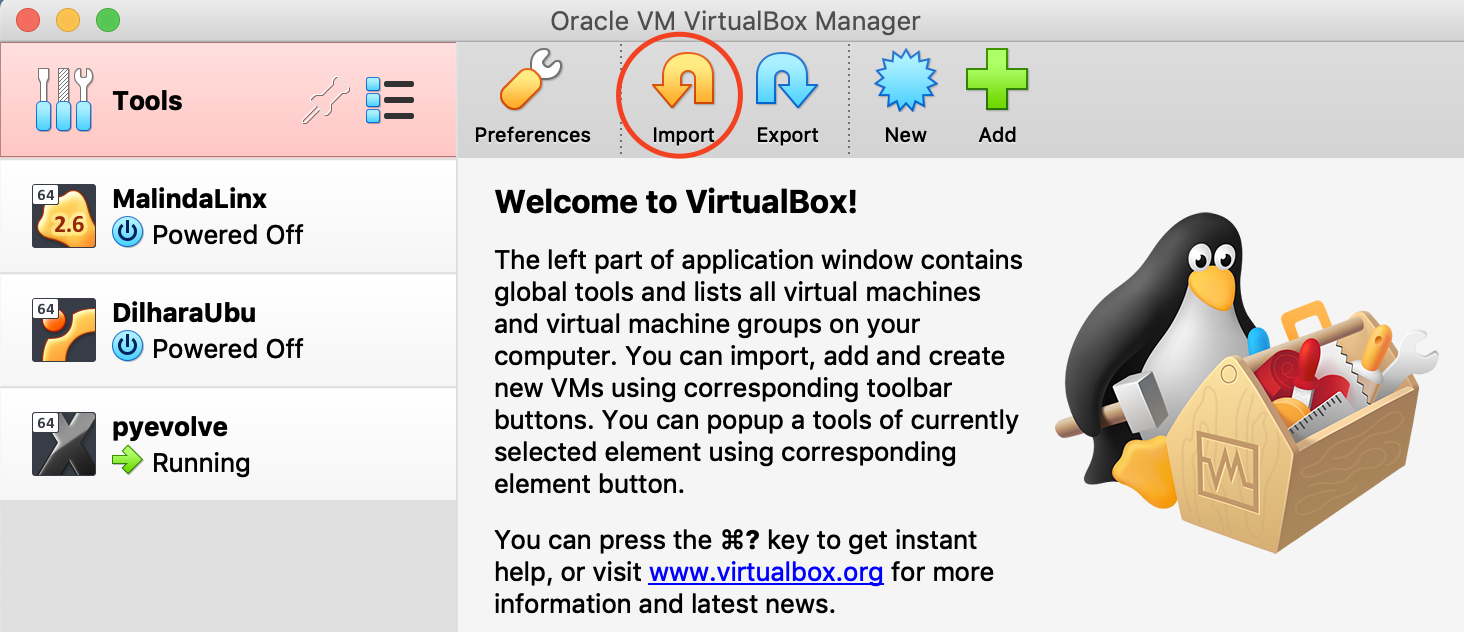
Step 1.3: Open VirtualBox. Click on the tools tab and then import the downloaded image file (pyevolve.ova) to VirtualBox, as shown below. For the image to be loaded into VirtualBox, you might need at least 40GB of free space. If not, you are likely to experience an error with the error code NS ERROR_INVALID_ARG.
You will be taken to the window shown below, where the settings should appear as depicted. Click the 'Import' button, as indicated in the image. This may take a few minutes to import the image into VirtualBox.
Step 1.4: To set up the configurations successfully, you need to disable the USB port of the virtual machine. To do this, follow the steps outlined in the following image. If this is not done properly, you will receive an error in the step Step 1.5 with the error code NS_ERROR_FAILURE.
disableUSB.mov
Step 1.5: Start the virtual machine by pressing the "Start" button at the top of the window.
Step 1.6: The virtual machine should ideally start now. However, sometimes it may enter into a shell prompt as shown below.
If this is the case, you will need to manually run the following two commands one after the other.
FS1:System\Library\CoreServices\boot.efi
The following video illustrates the steps described above (check Step 1.7: for login password).
load.mov
Step 1.7: The above step may take several minutes to start the machine, and you should see the startup screen as shown below. Use the password abc@123 to log in to the machine. Once logged in, you will find a folder named PYEVOLVE_FILES which contains the executables, source code, and data for PyEvolve.
You have successfully configured all the necessary setup for executing PyEvolve.
Under the evaluation, we demostrate transplantation of following patterns to the project Keras. Please refer the technical paper for the further information of the pattern.
\
:[[l1]] = open(:[[l2]], "r") ----\ with open(:[[l2]], "r") as :[[l1]]:
:[l4] = :[[l1]].readlines() ----/ :[[l4]] = :[[l1]].readlines()
:[[l1]].close() /These changes were submited to the project keras and was accepted throught this pull request (keras-team/keras#16874).
Step 2.1: Open the teminal application in the virtual machine and Navigate to the folder PYEVOLVE_FILES using the command cd ~/Desktop/PYEVOLVE_FILES.
Step 2.2: Execute ls to view the PyEvolve executable pyevolve-1.0-SNAPSHOT.jar and other data.
For your convenience, we have included all the commands needed in the following steps in the file ~/Desktop/commands.txt, so that you do not have to type the long commands.
Step 2.3: You can use the command java --enable-preview -jar pyevolve-1.0-SNAPSHOT.jar to view all the required input arguments to successfully run the tool (your working directory must be ~/Desktop/PYEVOLVE_FILES).
Below are the arguments for your knowledge (you do not have any action to perform).
-p,--patternsThis folder contains code change patterns with two types of filenames: those that begin with 'l_', and those that begin with 'r_'. These prefixes indicate the rule of a code change before and after. It is essential that the names following the prefixes are the same for the tool to correctly identify the files that belong to the same change. For example, you can check the folder~/Desktop/PYEVOLVE_FILES/PATTERNS/which contains patterns that we are planning to use in this evaluation.-r,--repositoriesThe path to the project repository where the code change must be transplanted. This folder houses all of the projects.-f,--filesThis file contains the project files that must be reviewed and modified. The paths has to be a relative path to the folder indicated by the argument-r.-t,--typesThis folder holds the "type" information of the program elements of the projects found in the project repository (-r). This information needs to be inferred and stored in this folder before running the "PyEvolve". Instructions for inferring type information can be found in this repository: https://github.com/mlcodepatterns/PythonTypeInformation. For your convenience, the type information has already been included in the VirtualBox Image.
Step 2.4: To clone the project keras to the folder ~/Desktop/PYEVOLVE_FILES/PROJECTS/keras-team/ please execute following commands.
- Navigate to the
cd ~/Desktop/PYEVOLVE_FILES/PROJECTS/keras-team/ - To clone
git clone https://github.com/keras-team/keras.git ./keras/ - To navigate to the folder
keras, executecd keras/ - To retrieve the previous snapshot, before merging the pull request, execute
git checkout f49e66c72ea5fe337c5292ee42f61cd75bc74727.
Step 2.5: To apply the patterns in the folder ./PATTERNS/ execute the following command. The argument descriptions are provided in the Step 2.3.
- You should first navigate to the folder
PYEVOLVE_FILESusing the commandcd ~/Desktop/PYEVOLVE_FILES/ - To apply patterns execute
java --enable-preview -jar pyevolve-1.0-SNAPSHOT.jar -r ./PROJECTS/ -f ./refactoring_files.txt -p ./PATTERNS/ -t ./TYPE_REPO/
The above command executes the main function of Pyevolve given in this link and makes the code changes for the project included in the folder ~/Desktop/PYEVOLVE_FILES/PROJECTS/.
Step 2.6: To check the changed files, navigate to the folder ~/Desktop/PYEVOLVE_FILES/PROJECTS/keras-team/keras execute git diff, scroll down to see all the changes. You can observe a successful transplantation of the patten given in Section 2.
You have successfully executed PyEvolve and transplanted patterns to the Keras project.
The steps 2.4, 2.5, and 2.6 are demonstrated in the video below.
evaluation4.mp4
We have made available two distinct types of large datasets. To evaluate PyEvolve, we utilized over 40,000 code transformation trials by utilizing a dataset of change patterns collected from actual repositories. This dataset is readily accessible to the public. Additionally, PyEvolve automatically transplanted code change patterns by submitting 40 pull requests to open-source repositories. We have also made the list of these pull requests publicly available.
PyEvolve was evaluated using the corss validation dataset. This data set contains frequent code changes made in open source Python projects. We evaluated PyEvolve with over 40,000 transformation trials. To provide easy access to data, we released data on a website hosted on GitHub, as described above. We have, however, archived all tools and data in accordance with ICSE 2023 Open Science Policies.
- Zendo link for cross validation dataset : https://zenodo.org/record/7566407#.Y9At3C1h2cY
| # | Created at | Url | State | Merged |
|---|---|---|---|---|
| 1 | 2022-06-26T18:21:33Z | HazyResearch/pdftotree#122 | closed | True |
| 2 | 2022-06-26T19:55:38Z | brightmart/text_classification#149 | closed | True |
| 3 | 2022-06-28T08:12:21Z | tensorflow/lattice#73 | closed | True |
| 4 | 2022-06-29T07:44:12Z | quadrismegistus/prosodic#37 | closed | True |
| 5 | 2022-07-01T01:31:54Z | idaholab/raven#1877 | closed | True |
| 6 | 2022-07-01T08:00:50Z | erikbern/ann-benchmarks#303 | closed | True |
| 7 | 2022-07-01T08:13:58Z | david-abel/simple_rl#61 | closed | True |
| 8 | 2022-07-04T09:41:28Z | microsoft/nni#4982 | closed | True |
| 9 | 2022-07-04T20:33:39Z | ray-project/ray#26284 | closed | True |
| 10 | 2022-07-05T00:03:01Z | jindongwang/transferlearning#341 | closed | True |
| 11 | 2022-07-06T06:48:04Z | dipy/dipy#2618 | closed | True |
| 12 | 2022-07-06T08:45:22Z | pgmpy/pgmpy#1551 | closed | True |
| 13 | 2022-07-14T08:44:44Z | reframe-hpc/reframe#2565 | closed | True |
| 14 | 2022-07-14T08:52:19Z | DeepLabCut/DeepLabCut#1905 | closed | True |
| 15 | 2022-08-06T06:16:59Z | pytorch/pytorch#82929 | closed | True |
| 16 | 2022-08-06T06:46:26Z | ray-project/ray#27600 | closed | True |
| 17 | 2022-08-06T08:04:47Z | keras-team/keras#16874 | closed | True |
| 18 | 2022-08-06T08:22:04Z | GoogleCloudDataproc/cloud-dataproc#152 | closed | True |
| 19 | 2022-08-06T08:29:03Z | facebookresearch/ParlAI#4718 | closed | True |
| 20 | 2022-08-06T09:35:13Z | idaholab/raven#1930 | closed | True |
| 21 | 2022-08-07T05:24:26Z | BindsNET/bindsnet#570 | closed | True |
| 22 | 2022-08-07T05:49:51Z | CellProfiler/CellProfiler#4610 | closed | True |
| 23 | 2022-08-07T06:32:24Z | daniellerch/aletheia#21 | closed | True |
| 24 | 2022-08-07T06:51:40Z | deepinsight/insightface#2070 | closed | True |
| 25 | 2022-08-06T07:12:28Z | scikit-image/scikit-image#6458 | open | True |
| 26 | 2022-08-07T06:14:46Z | danforthcenter/plantcv#932 | open | True |
| 27 | 2022-08-07T00:50:05Z | aws/sagemaker-python-sdk#3286 | closed | True |
| 28 | 2022-08-07T05:56:14Z | cesium-ml/cesium#309 | closed | True |
| 29 | 2022-07-05T01:40:15Z | LCAV/pyroomacoustics#271 | closed | True |
| 30 | 2022-06-27T02:46:23Z | Pinafore/qb#107 | open | False |
| 31 | 2022-06-29T02:33:54Z | tensorflow/transform#280 | open | False |
| 32 | 2022-06-29T05:18:34Z | tensorflow/ranking#325 | open | False |
| 33 | 2022-07-01T01:47:41Z | google-research/google-research#1189 | open | False |
| 34 | 2022-07-05T08:05:16Z | bnpy/bnpy#42 | open | False |
| 35 | 2022-07-05T08:23:56Z | brainiak/brainiak#516 | open | False |
| 36 | 2022-07-13T20:16:05Z | LxMLS/lxmls-toolkit#176 | open | False |
| 37 | 2022-06-26T22:09:38Z | cornellius-gp/gpytorch#2049 | closed | False |
| 38 | 2022-06-30T08:12:36Z | lmcinnes/pynndescent#192 | closed | False |
| 39 | 2022-07-05T05:41:14Z | pyRiemann/pyRiemann#185 | closed | False |
| 40 | 2022-07-11T06:56:15Z | calico/basenji#125 | closed | False |