This library is a Python implementation of random feature latent variable models (RFLVMs), which are a family of nonlinear dimension reduction methods that use random Fourier features to support non-Gaussian data likelihoods. RFLVMs were developed by Gregory Gundersen, Michael Zhang, and Barbara Engelhardt. See our paper for details.
This library was written by Gregory Gundersen. Please feel free to submit any bugs requests.
Gaussian process-based latent variable models are flexible and theoretically grounded tools for nonlinear dimension reduction, but generalizing to non-Gaussian data likelihoods within this nonlinear framework is statistically challenging. The main problem is that a non-Gaussian likelihood cannot be integrated out in closed form. Here, we use random features to develop a family of nonlinear dimension reduction models that are easily extensible to non-Gaussian data likelihoods; we call these random feature latent variables models (RFLVMs). By approximating a nonlinear relationship between the latent space and the observations with a function that is linear with respect to random features, we induce closed-form gradients of the posterior distribution with respect to the latent variable. This means we can use gradient-based methods, such as quasi-Newton methods or HMC, to optimize our posterior w.r.t. the latent variables. This allows the RFLVM framework to support computationally tractable nonlinear latent variable models for a variety of data likelihoods in the exponential family without specialized derivations.
To replicate results in the paper, call fit_model.py with appropriate arguments. The datasets bridges, congress, and s-curve are supported here. For example, to replicate the Poisson model, call
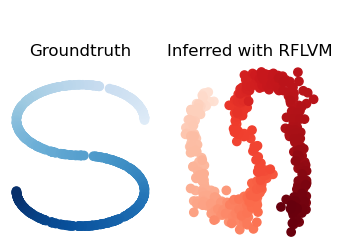
python fit_model.py --dataset=s-curve --emissions=poisson --model=poissonThis uses Scikit-learn's make_s_curve function to generate latent variables in the shape of an "S" and then generates data according to the data generating process described in the paper. To see the data generating process in code, see here. See the ArgumentParser instance in fit_model.py for a description of arguments.
This implementation requires Python 3.X. See requirements.txt for a list installed packages and their versions. The main packages are:
autograd
GPy
matplotlib
numpy
pandas
pypolyagamma
scipy
scikit-learn