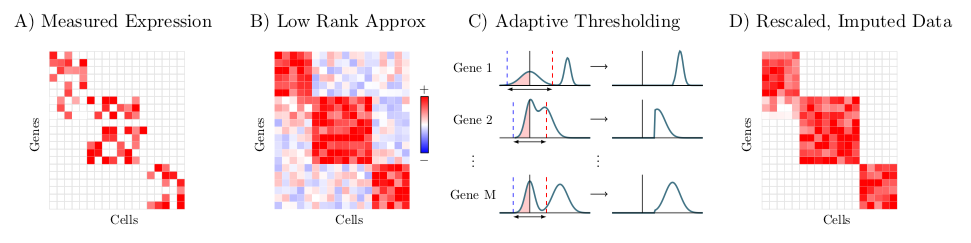
ALRA is a method for imputation of missing values in single cell RNA-sequencing data, described in the preprint, "Zero-preserving imputation of scRNA-seq data using low-rank approximation" available here. Given a scRNA-seq expression matrix, ALRA first computes its rank-k approximation using randomized SVD. Next, each row (gene) is thresholded by the magnitude of the most negative value of that gene. Finally, the matrix is rescaled.
This repository contains codes for running ALRA in R. The only prerequisite for ALRA is installation of the randomized SVD package RSVD which can be installed as install.packages('rsvd').
Please be sure to pass ALRA a matrix where the cells are rows and genes are columns.
ALRA can be used as follows:
# Let A_norm be a normalized expression matrix where cells are rows and genes are columns.
# We use library and log normalization, but other approaches may also work well.
result.completed <- alra(A_norm)
A_norm_completed <- result.completed[[3]]
See alra_test.R for a complete example.
ALRA is integrated into Seurat v3.0 (currently at pre-release stage) as function RunALRA(). But if you use Seurat v2, we provide a simple function to perform ALRA on a Seurat v2 object in alraSeurat2.R.