New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Reading large EDF files with preload = False raises memory error #10634
Comments
|
Hello! 👋 Thanks for opening your first issue here! ❤️ We will try to get back to you soon. 🚴🏽♂️ |
|
Would you be able to run Oh, and before that, does the problem also occur outside of a notebook (e.g. plain Python script or Python interactive interpreter, or even IPython)? |
|
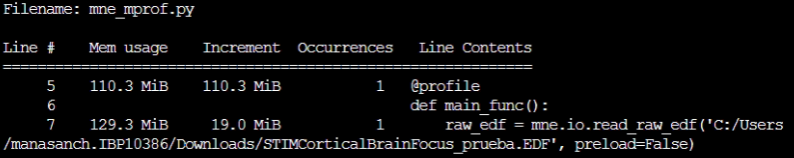
Thanks for the rapid response. Yes, it also occurs outside a notebook, both in plain Python script and in a Python interactive interpreter. I have run the The final increment is only 19Mib (the size of the metadata I assume), but the process to get there does not only require these 19Mib, but more than 10Gbs approx. Should we go deeper in the source code? Let me know if I can help with anything else. |
|
I fear you need to dig into the code and put breakpoint or using something
like memory_profiler to identify the pb
… Message ID: ***@***.***>
|
|
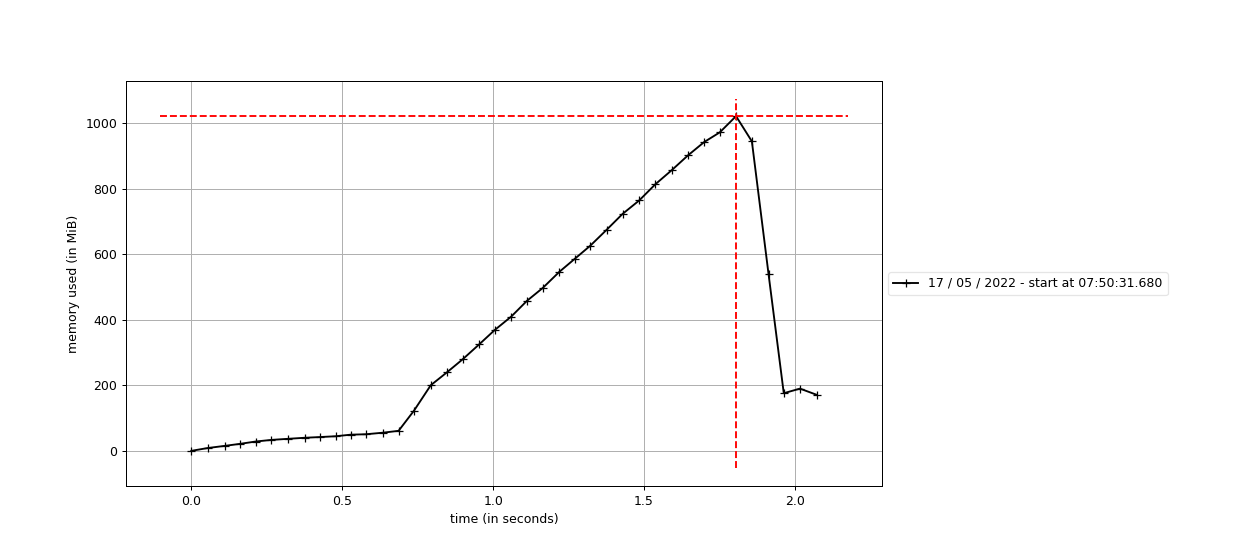
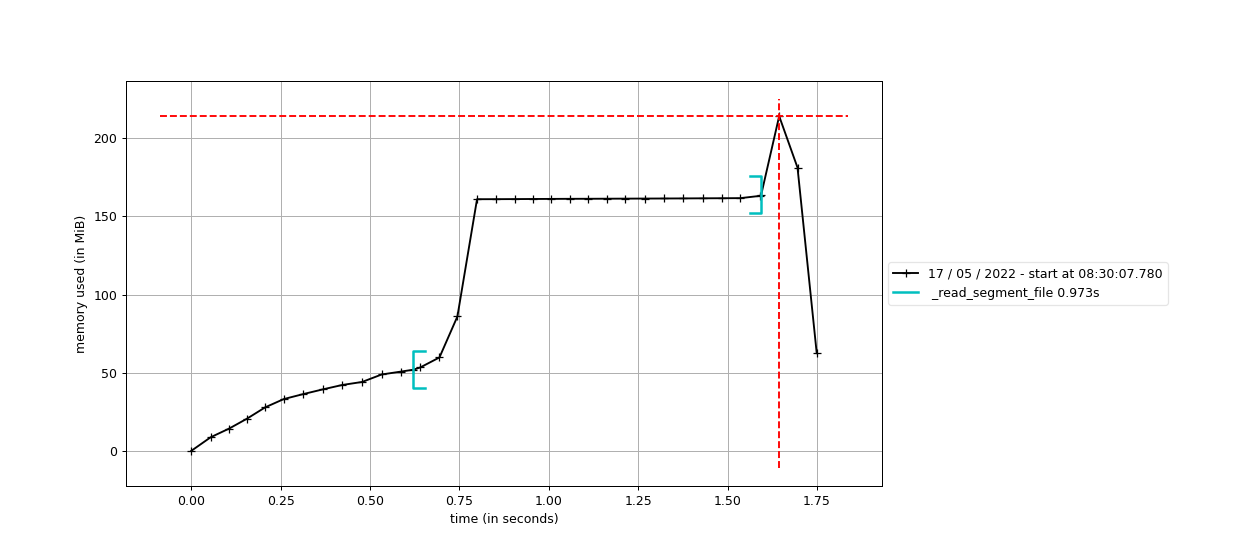
Can you do a time-based memory profile (https://github.com/pythonprofilers/memory_profiler#time-based-memory-usage)? This should at least show the 10GB spike at some point. |
|
Thanks! I guess the next thing to do would be to sprinkle some |
|
So, I executed the memory_profile over the source code and the RAM gets filled in the It is in the
If I keep track of the RAM before and after this line execution (using Let me know if you need any other information. Thanks for the help. |
|
Thanks @arnaumanasanch, I'll take a look to see why this is necessary without |
|
If anyone wants to reproduce the problem, here's a reprex that generates a large EDF file (944MB on disk, 3.6GB in RAM, but values can be adapted) and reads it with import numpy as np
from mne.io import read_raw_edf
from pyedflib.highlevel import write_edf_quick
def write_large_edf():
n_chans = 64
length = 2 * 60 * 60
fs = 1024
write_edf_quick("large.edf", np.random.randn(n_chans, length * fs), fs)
raw = read_raw_edf("large.edf", preload=False)Running that script with |
|
I think I found one place where we accidentally create a view on an array, which prevents garbage collection and therefore fills up memory. This line creates a reference to I'll submit a PR so that you can test with your file @arnaumanasanch. |


Issue/Bug
When reading in an IPython notebook a large .edf file which is 10Gb size (4 hours/ 150 channels/ 2048Hz sampling frequency) with:
raw_edf = mne.io.read_raw_edf('file.edf', preload=False)the kernel crashes as the RAM memory, which is 12 Gb, gets full.
I thought the preload argument was loading only metadata (which should not be more than a few hundred Mbs).
Would there be a way in which I could read the raw_edf (only the metadata) and then with the get_data() method, be able to just load into memory a small piece of data (n channels and x time range) without the system failing because of the RAM being full?
If I try the same with a higher RAM (32Gb), there is no problem and once it is loaded, the RAM goes back to normal (e.g: before loading it is 1Gb, when loading it goes up to 15Gb, once loaded back to 1 Gb approx.). We need to make it functional, if possible, with the 12 Gb RAM.
For privacy reasons, I can’t share the file that I am using.
Thanks in advance.
Additional information
MNE version: e.g. 1.0.3
operating system: / Windows 10
The text was updated successfully, but these errors were encountered: