Source code for my publication,<Analyzing angiogenesis on a chip using deep learning-based image processing> https://pubs.rsc.org/en/content/articlehtml/2023/lc/d2lc00983h
!! This code implemented the NuSet, https://github.com/yanglf1121/NuSeT For Nuclei segmentation process. I want to show my deepest appreciation to the authors of NuSeT, https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008193
Installation :
- Download files or clone this git.
- At your python terminal, please install all required modules by entering followings:
pip install -r requirements.txt
!Important!
This application is optimized for following versions of modules :
python : 3.7.0
tensorflow-gpu:1.15.0
numpy:1.19.0
If error occurs during installation, please refer the git of yanglf1121.
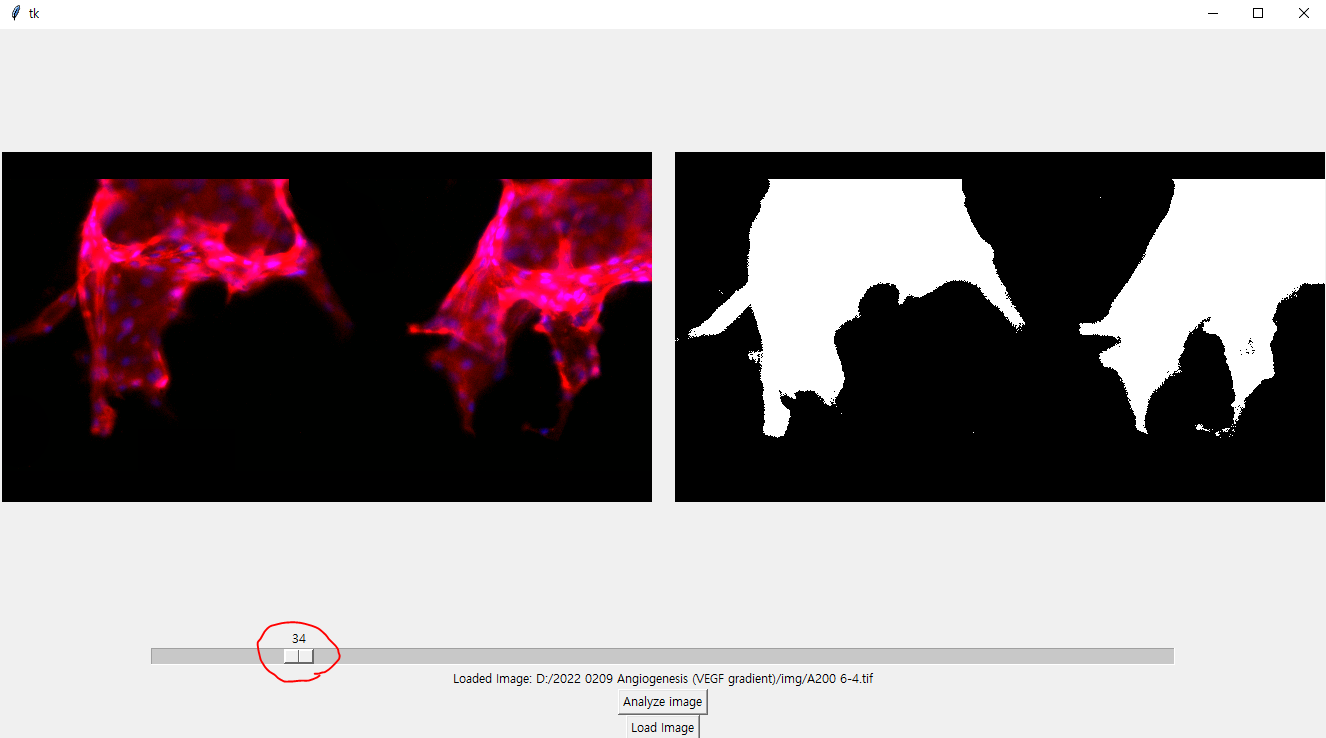
Also, PLEASE MAKE SURE THAT THE IMAGE YOU WANT TO ANALYZE HAS NUCLEI INFORMATION ON BLUE CHANNEL, AND VASCULAR INFORMATION ON GREEN AND RED CHANNEL.
- The preview offers upper-left part of your image. please take note for this.