-
-
Notifications
You must be signed in to change notification settings - Fork 0
sonar_tutorial

The original dataset contains 208 examples organized on 60 features:
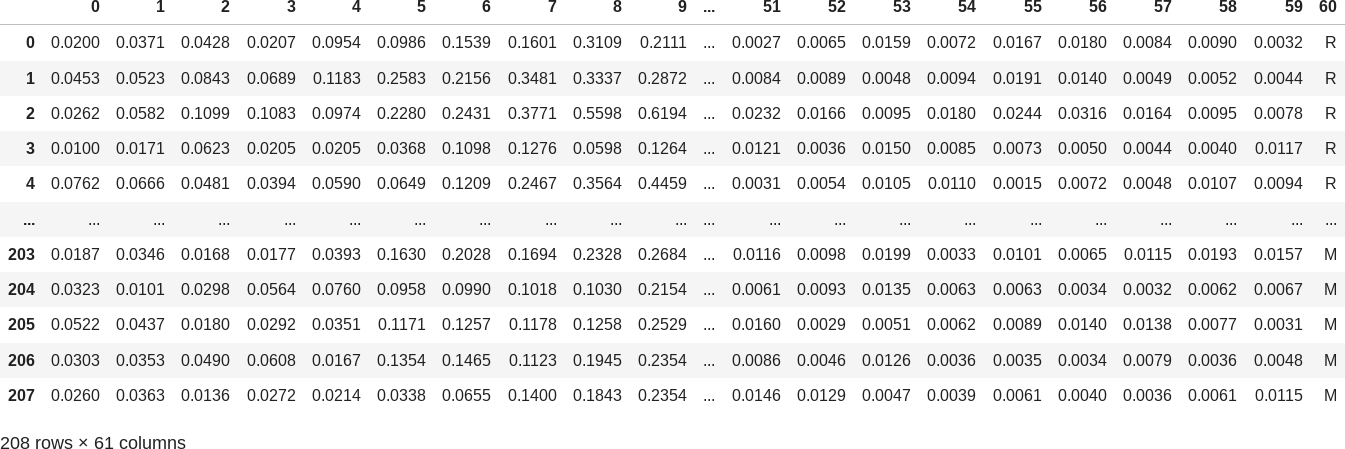 (you can take a look at sonar.md for additional information about the dataset)
(you can take a look at sonar.md for additional information about the dataset)
The last column holds the label / class of the examples (values "R" and "M", which stands for "Rock" or "Mine" observation). We will try to predict it based on the columns from 0 to 59.
We can look at the class balance:
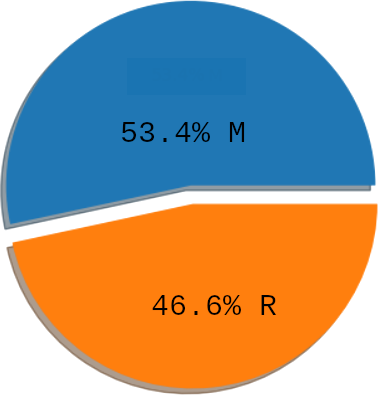
concluding that there isn't a class imbalance problem.
#include "kernel/ultra.h"
int main()
{
using namespace ultra;
// READING INPUT DATA
src::dataframe::params params;
params.output_index = src::dataframe::params::index::back;
src::problem prob("sonar.csv", params);
// ...
}Data preprocessing isn't required (take a look at dataset format for details):
- all attributes / features are numerical;
- there isn't a header row (usually it's automatically identified).
All we have to do is point the column containing labels:
params.output_index = src::dataframe::params::index::back;Actually there's just a little bit left before the first attempt so let's add the missing code without hesitation:
int main()
{
using namespace ultra;
// READING INPUT DATA
src::dataframe::params params;
params.output_index = src::dataframe::params::index::back;
src::problem prob("sonar.csv", params);
// SETTING UP SYMBOLS
prob.setup_symbols();
// SEARCHING
src::search s(prob);
s.validation_strategy<src::holdout_validation>(prob, 70);
const auto result(s.run());
std::cout << "\nCANDIDATE SOLUTION\n"
<< out::c_language << result.best_individual
<< "\n\nACCURACY\n" << *result.best_measurements.accuracy * 100.0
<< '%'
<< "\n\nFITNESS\n" << *result.best_measurements.fitness << '\n';
}All the classes and functions are placed into the ultra namespace.
Now compiling and executing the code (for your ease the above code is in the examples/sonar.cc file):
$ cd build/examples
$ ./sonaryou should see something like (actual values will differ):
[INFO] Importing dataset...
[INFO] ...dataset imported
[INFO] Examples: 208, features: 60, classes: 2
[INFO] Setting up terminals...
[INFO] Category 0 variables: `X1` `X2` `X3` `X4` `X5` `X6` `X7` `X8` `X9` `X10` `X11` `X12` `X13` `X14` `X15` `X16` `X17` `X18` `X19` `X20` `X21` `X22` `X23` `X24` `X25` `X26` `X27` `X28` `X29` `X30` `X31` `X32` `X33` `X34` `X35` `X36` `X37` `X38` `X39` `X40` `X41` `X42` `X43` `X44` `X45` `X46` `X47` `X48` `X49` `X50` `X51` `X52` `X53` `X54` `X55` `X56` `X57` `X58` `X59` `X60`
[INFO] ...terminals ready
[INFO] Automatically setting up symbol set...
[INFO] Category 0 symbols: `FABS` `FADD` `FDIV` `FLN` `FMUL` `FMOD` `FSUB`
[INFO] ...symbol set ready
[INFO] Holdout validation settings: 70% training (145), 15% validation (31), 15% test (32)
[INFO] Number of layers set to 1
[INFO] Population size set to 740
2.001 0: -51.308
6.004 0: -50.317
14.010 0: -46.4347
01:08 3: -45.186
01:14 3: -45.186 [1x 145gph]
At last we have a program to classify training examples! Not too difficult, but what's going on under the hood?
src::problem prob("sonar.csv", params);src::problem class is a specialization of the ultra::problem class for symbolic regression and classification tasks. The constructor reads/stores a dataset (file "sonar.csv") and other useful information.
Classification (and symbolic regression) tasks are tackled through genetic programming. In genetic programming every candidate solution / program needs an alphabet to be expressed (primitive set).
The command:
prob.setup_symbols();sets the default primitive set. Part of this set (the terminal set) is derived from the input data
[INFO] Setting up terminals...
[INFO] Category 0 variables: `X1` `X2` `X3` `X4` `X5` `X6` `X7` `X8` `X9` `X10` `X11` `X12` `X13` `X14` `X15` `X16` `X17` `X18` `X19` `X20` `X21` `X22` `X23` `X24` `X25` `X26` `X27` `X28` `X29` `X30` `X31` `X32` `X33` `X34` `X35` `X36` `X37` `X38` `X39` `X40` `X41` `X42` `X43` `X44` `X45` `X46` `X47` `X48` `X49` `X50` `X51` `X52` `X53` `X54` `X55` `X56` `X57` `X58` `X59` `X60`
[INFO] ...terminals ready
and part (the function set) is arbitrarily chosen:
[INFO] Automatically setting up symbol set...
[INFO] Category 0 symbols: `FABS` `FADD` `FDIV` `FLN` `FMUL` `FMOD` `FSUB`
[INFO] ...symbol set ready
src::search s(prob);The search of solutions is entirely driven by the master src::search class. It uses prob to access data and specific parameters/constraints.
src::search is a template class, its template parameters allow to choose various search algorithms. For now we stick with the default.
Training and testing a model on the same dataset can lead to overfitting, where the model memorizes the training data rather than learning underlying patterns, resulting in poor generalization to new, unseen data.
For this reason available data are split into a:
- training set used for training a model;
- validation set to be able to measure the model generalization (i.e. to provide an unbiased evaluation of the model fit).
(see Training, validation, and test data sets for a good, simple introduction to the topic)
The instruction:
s.validation_strategy<src::holdout_validation>(prob);perform the split (and, by default, also shuffling and stratified sampling); hence the output message:
[INFO] Holdout validation settings: 70% training (144), 30% validation (64), 0% test (0)
const auto result(s.run());The run method executes the search and returns a summary including the best program found. Since we do not specify any environment parameter (e.g. population size, number of runs...) they're automatically tuned (e.g. see the information lines [INFO] Number of layers set to 1 and [INFO] Population size set to 740).
std::cout << "\nCANDIDATE SOLUTION\n"
<< out::c_language << result.best_individual
<< "\n\nACCURACY\n" << *result.best_measurements.accuracy * 100.0
<< '%'
<< "\n\nFITNESS\n" << *result.best_measurements.fitness << '\n';The interesting part is the accuracy that is calculated on the validation set. The candidate solution / individual isn't so useful: it acts as a discriminant function and so it's just a part of the classification model.
