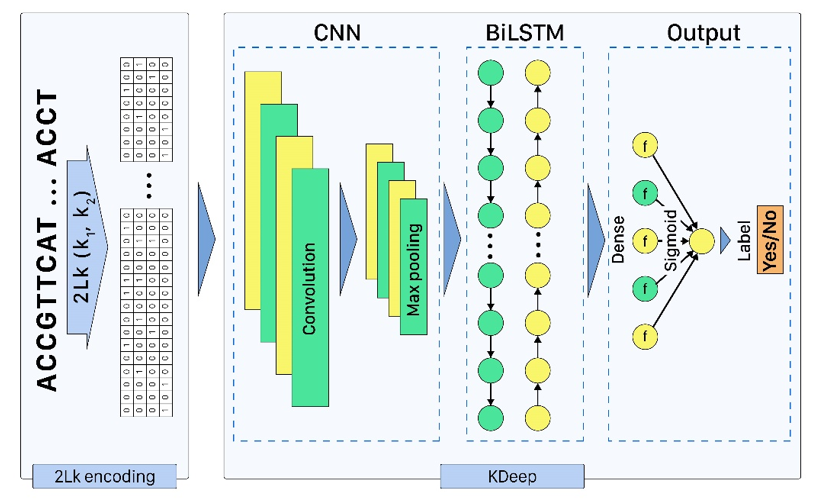
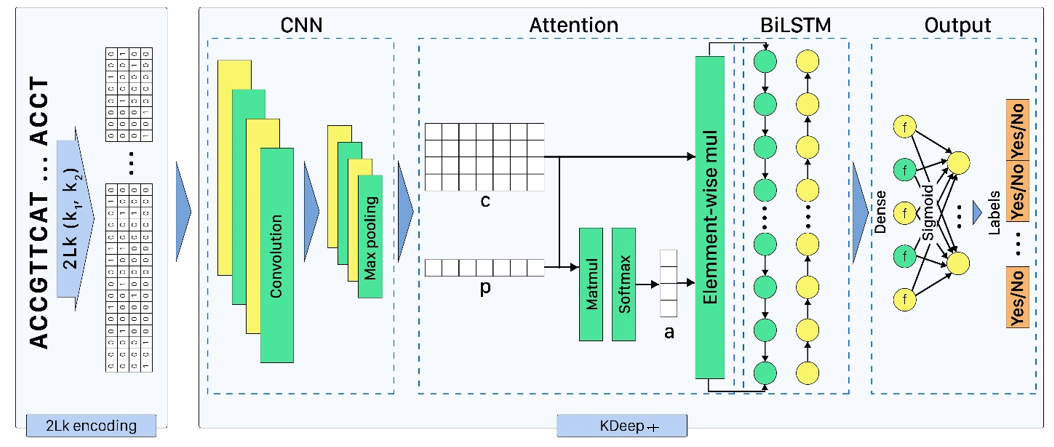
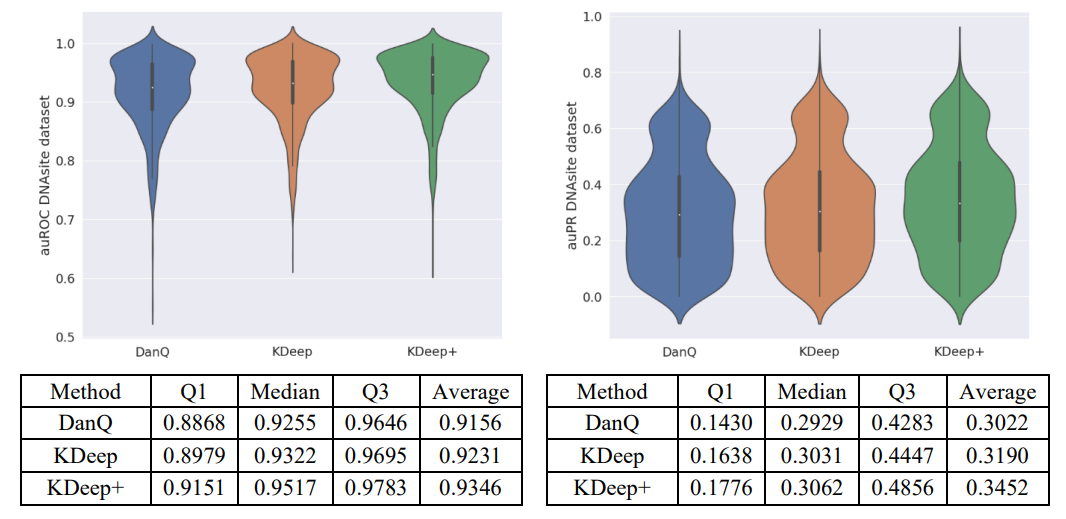
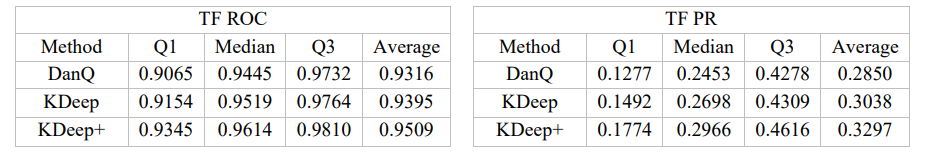
KDeep: a k-mer-based deep learning approach for predicting DNA/RNA transcription factor binding sites
Based on the importance of DNA/RNA binding proteins in different cellular processes, finding binding sites of them play crucial role in many applications, like designing drug/vaccine, designing protein, and cancer control. Many studies target this issue and try to improve the prediction accuracy with three strategies: complex neural-network structures, various types of inputs, and ML methods to extract input features. But due to the growing volume of sequences, these methods face serious processing challenges. So, this paper presents KDeep, based on CNN-LSTM and the primary form of DNA/RNA sequences as input. As the key feature improving the prediction accuracy, we propose a new encoding method, 2Lk, which includes two levels of k-mer encoding. 2Lk not only increases the prediction accuracy of RNA/DNA binding sites, but also, reduces the encoding memory-consumption by maximum 84%, improves the number of trainable parameters, and increases the interpretability of KDeep by about 79%, compared to the state-of-the-art methods.
python3.7, tensorflow==2.8, cuda and cuDNN if you have GPU
To train the model, download the training, validation and testing sets from DeepSEA dataset (You can download the datasets from here) After you have extracted the contents of the tar.gz file, move the 3 .mat files into the KDeep/data/ or KDeep+/data/ folder. then run below command:
1.python preprocess_FCGR.py. 2.python KDeep.py | KDeep+.py. 3.python test.py.
Skip download data from deepsea link. You need just download test data from here and here then extract files and move to DNA\KDeep\data or DNA\KDeep+\data folder. and download The KDeep model that trained by myself from here or KDeep+ model from here and move to DNA\KDeep\model or DNA\KDeep+\model folder.
If you want just test KDeep without training go to colab.
If you want just test KDeep+ without training go to colab.
Download Datasets from RNA_31then move to RNA\RNA_31 folder. AND RNA_24 then move to RNA\RNA_24 folder.
go to colab and run codes step by step.
For RNA-31:
python PreProcess.py
- Enter your direction of experience_train like (RNA_31/train/1/sequences.fa)
- Enter your direction of experience_test like (RNA_31/test/1/sequences.fa)
- Enter (fasta) to determine type of your data
For RNA-24:
python PreProcess.py
- Enter your direction of experience_train like (RNA_24/1/ALKBH5_Baltz2012_train)
- Enter your direction of experience_test like (RNA_24/1/ALKBH5_Baltz2012_test)
- Enter (text) to determine type of your data
For RNA-31: pythin Training.py
- Enter (420) to determine appropriate seed for learning
- Enter train number =(30000)
- Enter valid number = (10000)
- Enter batch_size = (300)
- Enter 101 to determine sequences lenght of RNA-31
For RNA-24: pythin Training.py
- Enter (0) to determine appropriate seed for learning
- Enter train number =(Check output of preprocess section) for experience one 'ALKBH5_Baltz2012' training number is 2410
- Enter valid number = (Check output of preprocess section). for experience one 'ALKBH5_Baltz2012' valid number is 266
- Enter batch_size like (300)
- Enter 375 to determine sequences lenght of RNA-24 Point=If the model fails to train, you should reduce the batch number
For RNA-31:
python Test.py
- Enter your direction of experience_test like (RNA_31/test/1/sequences.fa)
- Enter (fasta) to determine type of your data
- Enter (101) to determine sequences lenght of RNA-31
For RNA-24:
python Test.py
- Enter your direction of experience_test like (RNA_24/1/ALKBH5_Baltz2012_test)
- Enter (text) to determine type of your data
- Enter (375) to determine sequences lenght of RNA-24
For RNA-31:
python Training.py
- Enter your direction of experience_test like (RNA_31/test/1/sequences.fa)
- Enter (fasta) to determine type of your data
- Enter batch-size that use in trainin section
For RNA-24:
pyhton Training.py
- Enter your direction of experience_test like (RNA_24/1/ALKBH5_Baltz2012_test)
- Enter (text) to determine type of your data
- Enter batch-size that use in trainin section
Somayyeh Koohi
Department of Computer Engineering
Sharif University of Technology
e-mail: koohi@sharfi.edu