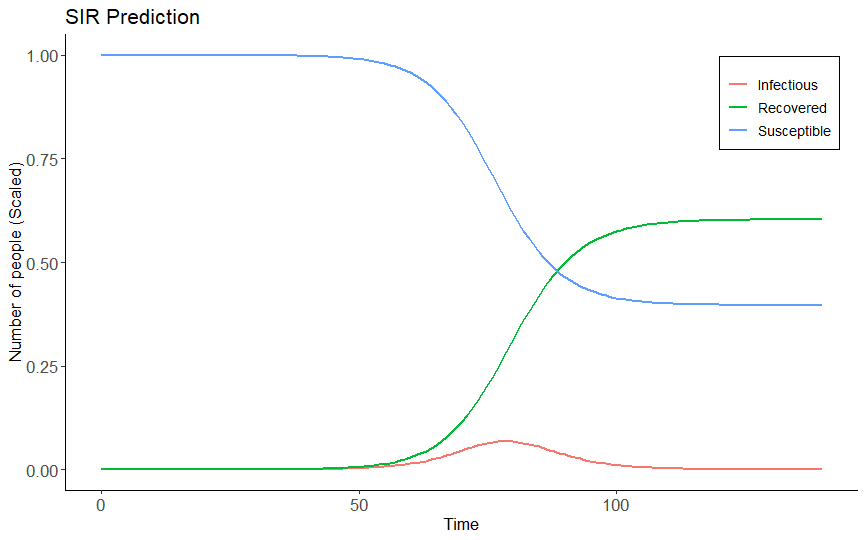
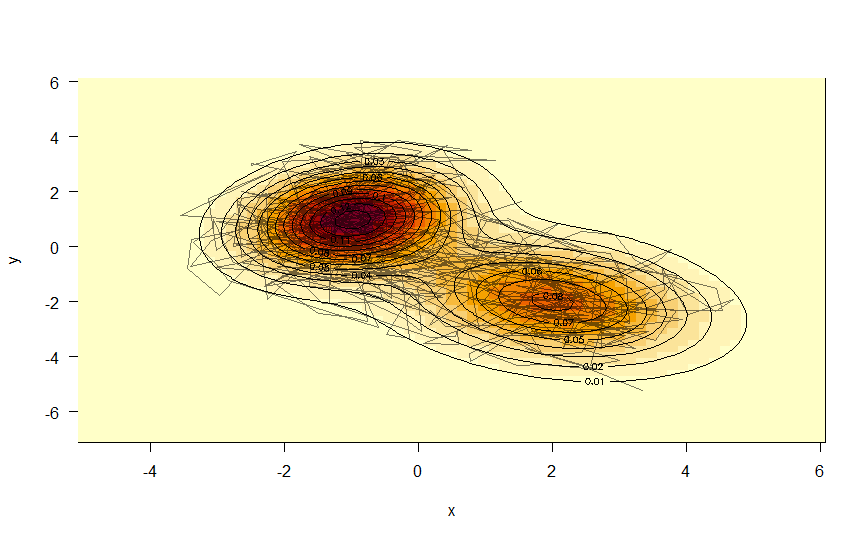
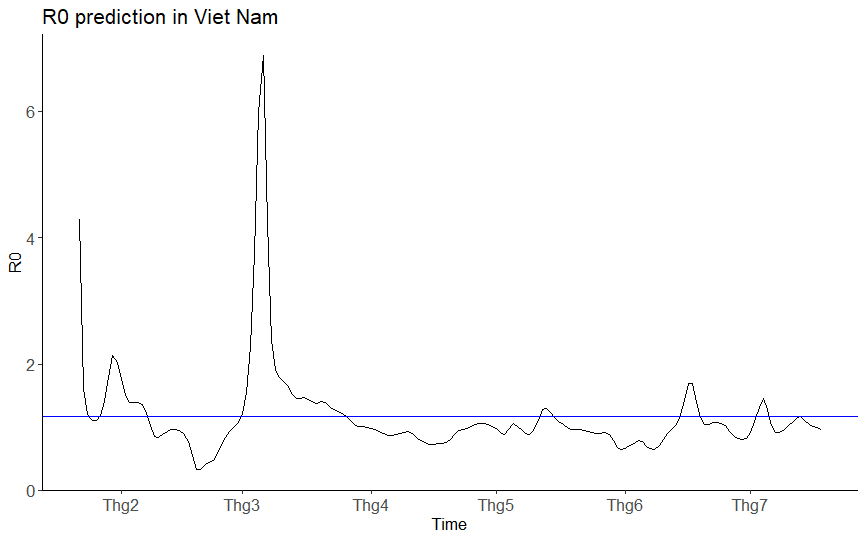
A project contains simple programs written in R language to implement SIR Model - a compartmental model for modeling infectious diseases mathematically.
More detailed information about the SIR Model can be found in here.
First, install R from CRAN and (optional) RStudio from here (You can use any IDE which supports R. I will take RStudio as a recommendataion).
Then, download the files inside source-code, open and run it by RStudio!
There are some packages I have used in this project. You can install these packages directy from RStudio by typing below line in RStudio terminal.
install.packages('package_name')For remoted packages derived from GitHub, first install devtools package
install.packages('devtools')and then install them by typing below line in RStudio terminal.
remotes::install_github("package_github_path")There are three programs in this project:
eulerMethod.R: A program to calculate the results of discrete SIR Model by using Euler method for solving first order ordinary differential equation.MCMC.R: Implement Random-Walk Metropolis-Hastings algorithm to generate samples of unknown probability distributions.estimateR0: Use real-time data and external libraries to predict the basic production ratio (R0) of a diseases in a specific country.
There are two tests inside the code of MCMC.R and estimateR0. To run them, just comment out the snippet of another test and run the program.
This project could not be done without the help from my lecturer in HCMC University of Technology, Dr. Nguyen An Khuong. This is also my individual project of his Mathematical Modeling course.
To complete the report about this project, I have used these papers as references:
- Kermack, W. O., & McKendrick, A. G. (1927). A contribution to the mathematical theory of epidemics. Proceedings of the royal society of london. Series A, Containing papers of a mathematical and physical character, 115 (772), 700-721.
- Bohner, M., Streipert, S., & Torres, D. F. (2019). Exact solution to a dynamic SIR model. Nonlinear Analysis: Hybrid Systems, 32, 228-238.
- Brauer, F. (2008). Compartmental models in epidemiology. In Mathematical epidemiology (pp. 19-79). Springer, Berlin, Heidelberg.