A simple and friendly Python framework for genetic-based algorithms (strongly supports tree-encoding)
- Supported algorithms: Genetic Algorithm (
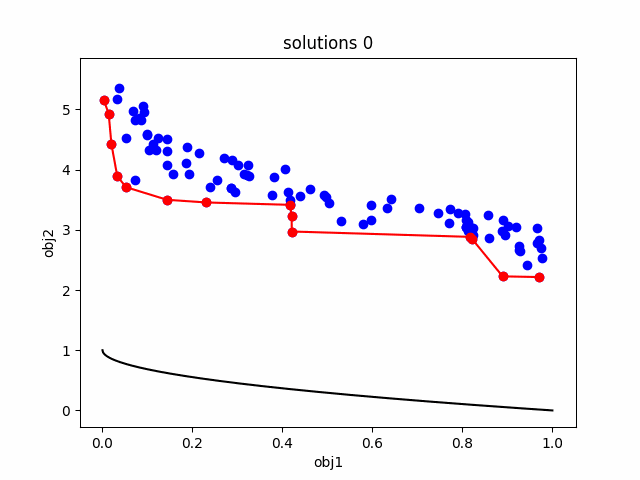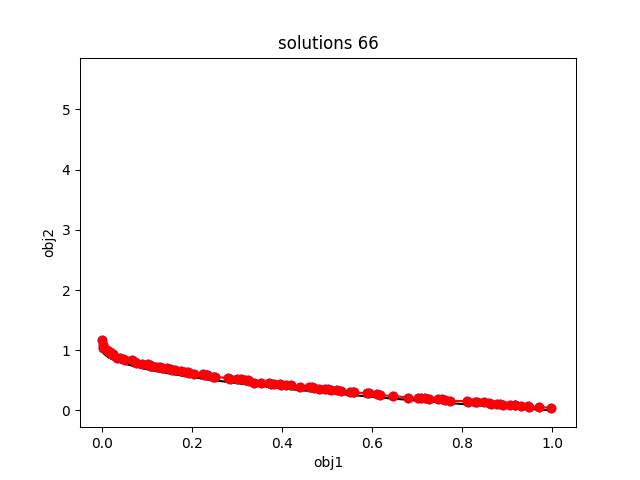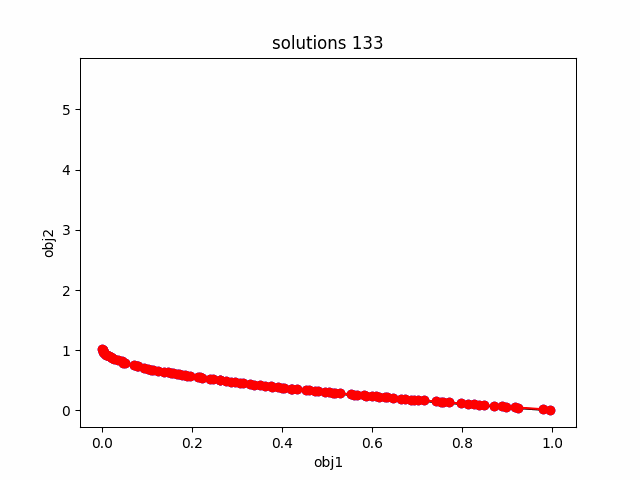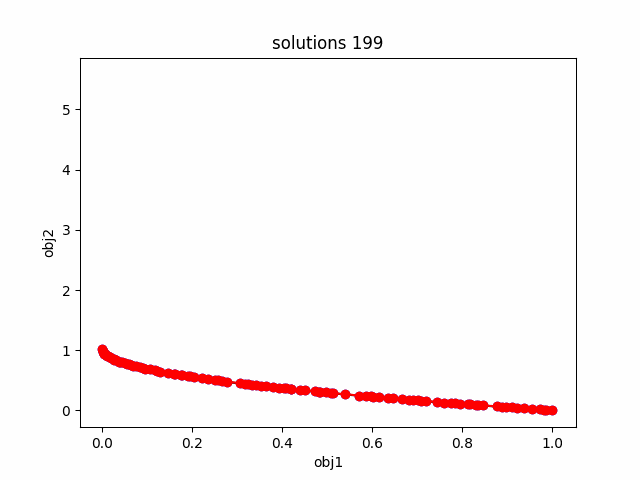
GAEngine), NSGA-ii (NSGAIIEngine). - An example on ZDT1 problem:
This package requires python 3.6 or later.
pip install geneticpython
We can quickly design a genetic algorithm in the following steps:
- define a individual template with specific encoding
from geneticpython.models import BinaryIndividual
indv_temp = BinaryIndividual(length=100)- define population based on created individual template. This population can uniformly initialize a population or you can define your own by passing
init_populationargument function
from geneticpython import Population
population = Population(indv_temp, pop_size=100)- define some core operators in genetic algorithm
from geneticpython.core.operators import RouletteWheelSelection, UniformCrossover, \
FlipBitMutation, RouletteWheelReplacement
selection = RouletteWheelSelection()
crossover = UniformCrossover(pc=0.8, pe=0.5)
mutation = FlipBitMutation(pm=0.1)
# this function decides which individuals will be survived
replacement = RouletteWheelReplacement()- create an engine and register the defined population and operators
from geneticpython import GAEngine
engine = GAEngine(population, selection=selection,
selection_size=100,
crossover=crossover,
mutation=mutation,
replacement=replacement)- register fitness function which gets an individual and returns its fitness value
@engine.maximize_objective
def fitness(indv):
return fitness_of_indv- run engine
engine.create_seed(seed)
history = engine.run(generations=1000)- get results and plot history
ans = engine.get_best_indv()
print(ans)
plot_single_objective_history({'geneticpython': history})You can find more examples here
This project is in development, if you find any issues, please create an issue here.
- Create extensive documentation and docs and comments in source-code
- Implement other algorithms:
PSO, DE, MOED/A, MOPSO, MODE,... - Implement other operators:
PMX crossover, ... - Create unit tests.
The goal of this project is to be able to build a simple and novice-friendly library yet functional enough to experiment with research projects. It is spontaneous and non-profit and also flawed.
We appreciate all contributions. If you are interested in contributing this project (including functional implementation or standard examples), please check Contribution page.
If you plan to contribute new features, utility functions, fix bugs, or extensions to the core, please first open an issue and discuss the feature with us.
Ngoc Bui (ngocjr7)
Special thanks to https://github.com/PytLab/gaft for getting me started a great API design.
This repository includes some parts of the following repos: