New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ND2: Incorrect dimensions #3502
Comments
|
A further example of this issue has been reported in forum thread https://forum.image.sc/t/hyperstack-ordering/34041 A further sample file was provided in QA-29075 and the bug was reproduced using Bio-Formats 6.3.1 In this case the ordering of the positions is incorrect and results in an ordering similar to the below: |
|
This issue has been mentioned on Image.sc Forum. There might be relevant details there: https://forum.image.sc/t/nd2-metadata-and-import-issues/40699/2 |
|
@dgault Hi David, |
|
@Parm0n some of our sample ND2 files used for the non-regression builds are available publicly here. IDR has also published studies in this format e.g. idr0095 or idr0109 and the raw data can be downloaded using Aspera. |
|
This issue has been mentioned on Image.sc Forum. There might be relevant details there: https://forum.image.sc/t/nd2-video-frame-time-stamp-corruption-in-imagej-metadata/38012/32 |
|
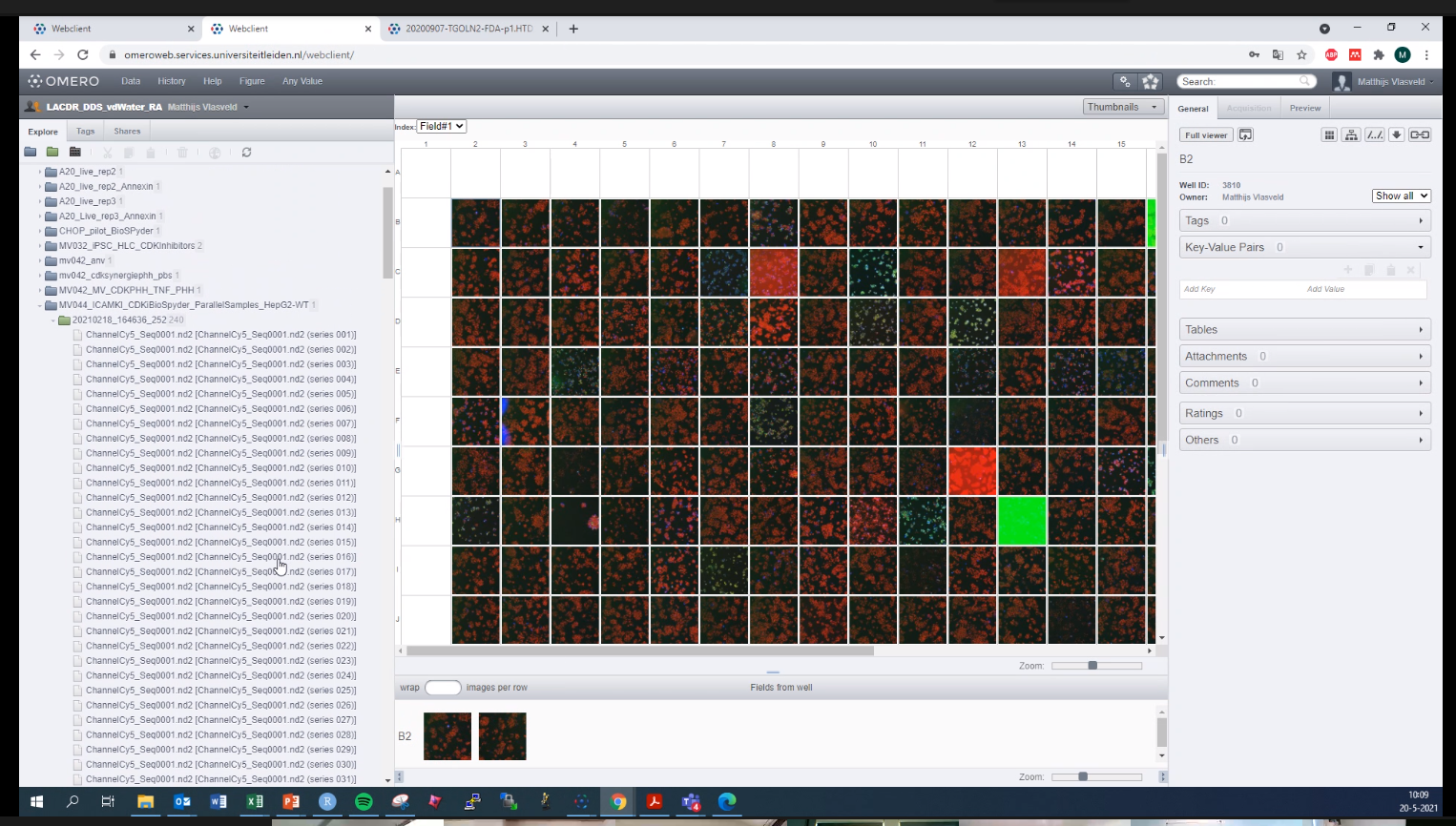
Hi @sbesson , Thanks for sharing the ND2 files. I think we were able to solve the problem related to incorrectly read positions and timepoints, but I'm not completely sure that this was the bug that is being solved in this conversion :) . Could you please provide access, if possible, to the files that are causing the import problem for you, so that I can test them. Hi @dgault, I have a question for you regarding the OMERO.web application, which may also be related to the problem discussed here. I have a screenshot of the OMERO.web application, which shows that some .nd2 document is open. This file imaged as a grid in which the parts of this document are located, in addition, below you can see a separate view for the fields from well. I also found this on the application documentation page (https://omero-guides.readthedocs.io/en/latest/introduction/docs/data-management.html Step-by-Step -> Browsing and rendering -> Point number 9). The question is how can I see this interface for well plates? Thank you :) |
|
Support for Screens and Plates is on a format by format basis and currently is not supported for ND2 data. If you want to check if it is supported for a specific format you should be able to see via the supported metadata page for the format. ND2: https://docs.openmicroscopy.org/bio-formats/6.6.1/metadata/NativeND2Reader.html for example compared to Perkin Elmer Columbus which supports Screen and Well metadata: https://docs.openmicroscopy.org/bio-formats/6.6.1/metadata/ColumbusReader.html |
|
Hi dgault, we are aware that support for well plates is currently missing for nd2, but we would really like to change this. The information from nd2 side is there as we have well plate views within NIS elements, we just need to make it available to you. Can you help us with an advise for the best way how to do this? Thanks a lot for your help :) |
|
Hi @SimoneNikon, thank you for reaching out. If you are able to link to or provide an existing specification or documentation on how to identify the relevant metadata within the format that would be a great help. If you would need to share privately you can submit it using https://www.openmicroscopy.org/qa2/qa/upload/. Alternatively if developers wish to open a PR to add support for plate views I can help point to the correct API calls required, a good example of how to populate the required metadata would be https://github.com/ome/bio-formats-examples/blob/master/src/main/java/FileExportSPW.java#L166 or https://github.com/ome/bioformats/blob/develop/components/formats-gpl/src/loci/formats/in/MetamorphReader.java#L861-L893 |
Issue was raised in the thread https://forum.image.sc/t/bioformats-reads-nd2-file-series-incorrectly/33263 and a sample file is provided in QA-29035
I have been able to reproduce the issue as described with Bio-Formats 6.3.1
The incorrect planes can be spotted by comparing the planes with the correct ordering in NIS Viewer. None of the ND2 configuration options have any impact on the result.
The text was updated successfully, but these errors were encountered: