New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Investigate slow OpenCL performance on AMD #3937
Comments
That's a mystery. There shouldn't be anything happening in between. No data transfer, no synchronization. We just launch one kernel, then launch the other.
In between those we do an
This is where we wait on that event. But that doesn't make sense as the explanation. We already had one long pause, and transferring a few bytes from device to host shouldn't take hundreds of microseconds. |
|
There is one way to quickly test the theory. Perform multiple instances of the simulation in parallel, utilizing multiple CPU cores. I hypothesize that running two simulations will double the throughput in units of @peastman I suspect this is harming performance on GPUs besides AMD. You or I could test the hypothesis on an M1 GPU. |
|
It's clear that the gaps are hurting performance. The question is, what causes them? |
|
How about we narrow down the problem? For example, remove one of the kernels that isn't causing a gap. See whether the gap persists. Repeat the procedure, you get the idea. This will narrow down and validate that, in isolation, e.g. |
|
I commented out the void OpenCLNonbondedUtilities::computeInteractions(int forceGroups, bool includeForces, bool includeEnergy) {
...
//downloadCountEvent.wait()
//updateNeighborListSize();
}The gap between |
You commented out the part that rebuilds nearest neighbor lists, so this is misleading. |
Sadly, I don't think that's real. It just means you broke it! We don't know in advance how much memory we'll need for the neighbor list. We pick an amount that we hope will be enough, but often isn't. Since the GBSA benchmark uses a larger cutoff than the others, it isn't. That's ok. The nonbonded kernel spots that there wasn't enough memory for the neighbor list and returns without doing much.
That only happens on Windows. It was added for Folding@home. Previously it had a tendency to load the GPU so heavily that the computer became unresponsive and people couldn't do anything else while it was running. We added the flush to make it break and ensure other processes would have time to run. But only on Windows, since on other platforms it probably isn't running as part of Folding@home. |
|
I suggested a better way to examine the problem, run two simulations in parallel. This requires a more complex setup than Benchmarks on M1 Max with the Metal plugin. Some benchmarks show significant underutilization, even with correct SIMD width. Remember that before, apoa1rf was using 13 W.
|
|
I know I broke the simulation...but in the process I think I confirmed |
|
The PME benchmark is a better test for this. It never needs to resize the neighbor list, so we can safely remove the related calls and see what difference it makes. Here's what I get. (I finally got my AMD GPU working again!) Original: 320 ns/day The download causes about a 25% slowdown, even though it's supposed to happen asynchronously and nothing ever depends on it having completed. |
|
|
My results are similar: Original: 300 ns/day
Can this be known ahead of time in more situations? If so, wrapping the |
Is this also the case in Reaction Field, the primary benchmark I'm measuring progress with?
Before doing this, we should figure out what's happening with AMOEBA PME. |
|
Unfortunately, it can never be known ahead of time. It also monitors the number of interactions for clues when it should reorder the molecules. That's important for performance in certain situations. |
|
@egallicc would you mind comparing power consumption to ns/day on HIP like I did, but without erasing |
|
Are you sure that the gap is not caused by |
|
Is this the data transfer from GPU -> CPU? Perhaps it's avoidable using indirect command buffers in Metal, HIP, CUDA, OpenCL 2.0. Have the GPU encode any commands depending on the value of Also how large is the buffer? It might take significant time over PCIe. If this hypothesis is correct, the gap you highlighted won't appear on M1 Pro. Also we could profile how long it takes the CPU to encode kernels (very expensive). |
1 uint What happens if |
|
OpenCL is encoding the findInteractionsWithBlocks kernel, issuing an event right after, then immediately requesting everything to finish. You're stalling the command stream just to read one value. With Metal that would invoke a 150+ us penalty. |
|
What do you mean by "encoding"? |
|
I mean issuing the command, making calls into a GPGPU API that the GPU will then execute. I think my analysis was slightly wrong; the OpenCL CPU-side code encodes the force kernel before waiting. That shouldn't starve the GPU unless the driver refuses to flush cached-up commands, when you ask it to wait on an event. We could try forcing a |
|
Perhaps you could move this entire function onto the GPU (it seems parallelizable). Have the GPU write certain buffers, which it reads in another kernel. You have persistent GPU threads so perhaps you don't need advanced indirect dispatch API to specify dispatch params. openmm/platforms/common/src/ComputeContext.cpp Lines 488 to 655 in a056d5a
This removes the need to break the command stream. If you don't want to find interactions, simply return early inside the appropriate kernel. |
I will try but not before the weekend. Teaching has resumed. :( I wouldn't want to be the bottleneck on this. Would it help if I shared my docker image to build OpenMM with HIP on Linux? |
I don't have HIP - I have an Apple GPU but I meant "compare changes in the same style I did". The change is using OpenCL vs using HIP. Maybe someone else with AMD would install Linux? |
|
I found a solution. The following boosted apoa1rf from 115 ns/day to 129 ns/day. void MetalNonbondedUtilities::computeInteractions(int forceGroups, bool includeForces, bool includeEnergy) {
if ((forceGroups&groupFlags) == 0)
return;
KernelSet& kernels = groupKernels[forceGroups];
if (kernels.hasForces) {
// ADD THIS LINE TO EXISTING CODE
if (useCutoff && numTiles > 0)
context.getQueue().flush();
cl::Kernel& kernel = (includeForces ? (includeEnergy ? kernels.forceEnergyKernel : kernels.forceKernel) : kernels.energyKernel);
if (*reinterpret_cast<cl_kernel*>(&kernel) == NULL)
kernel = createInteractionKernel(kernels.source, parameters, arguments, true, true, forceGroups, includeForces, includeEnergy);
if (useCutoff)
setPeriodicBoxArgs(context, kernel, 9);
context.executeKernel(kernel, numForceThreadBlocks*forceThreadBlockSize, forceThreadBlockSize);
}
if (useCutoff && numTiles > 0) {
// ADD THIS LINE TO EXISTING CODE
if (kernels.hasForces)
context.getQueue().flush();
downloadCountEvent.wait();
updateNeighborListSize();
}
}
|
|
Interesting! I have no idea why that would make a difference. Here's what I see running the PME benchmark on a few different GPUs. On each one I tried four versions: the original code, removing the call to RTX 4080: It mostly makes no difference. Almost all versions are a steady 1424 ns/day. The only exception is the version where I removed the download, which was slightly slower but also seemed rather noisy. I suspect those runs were slower for an unrelated reason. M1 Pro: The original was 130 ns/day. All the others were 147. The slowdown comes from the Radeon RX 5700 XT: The original is 320 ns/day. As posted above, skipping the wait speeds it up a little to 340, and skipping the download speeds it up a lot to 406. Adding It looks like we may have a good optimization here. I just wish I understood why it worked! It feels a little like black magic. |
I would say that SYCL is the first universal, coordinated attempt to solve this problem. Khronos finally got it right, unlike with OpenCL. The issue - such a solution requires so much engineering effort, it's still in its infancy. Up until recently only Intel sponsored it, so it got lopped in with oneAPI. Most people think SYCL is just "another CUDA", "another HIP" but that's inaccurate. |
|
Keep in mind that our resources are very limited. As in, I'm the only developer who works full time on OpenMM! I'm also the only developer who has been around for anything close to the full life of the project. It's not just about how to get the fastest performance on a particular GPU right now. It's about how to create a codebase that's easy to maintain and easy to add new features to. It's also about building on technologies that are around for the long term. And it's about how to prioritize effort to best serve our users. Here's the reality we have to work with: the vast majority of production simulations are run on NVIDIA GPUs. That makes them more important than all other platforms put together. That's not a reality I'm very happy with. I want there to be more competition. I really want AMD to get serious about HPC. But I also can't afford to dedicate too much time to it, and I certainly can't do anything that would add a major ongoing maintenance burden. Writing a new platform is a lot of work, but maintaining it long term is much more work. I've also been burned by AMD repeatedly. The very first implementation of OpenMM ran on AMD GPUs. It was written in Brook+, which was the language they were promoting for GPGPU at the time. Then they abandoned it and switched to OpenCL, which they declared was the hot new thing they were 100% committed to. And they were... right until they suddenly declared that HIP was the new hot new thing they were 100% committed to. But maybe we should instead be using SYCL, which is the even newer new hot new thing. Through all of this, AMD has never shown any sign of really being committed to HPC, or even of understanding it. Example: their compute profiler doesn't even work on Linux. Example: instead of continuing to invest in OpenCL, a cross platform standard that lots of developers have put a lot of work into supporting, they decide the whole community needs to rewrite their code to support a new proprietary API. Sorry for complaining. :) It's hard not to be bitter when you've been through too many rounds of this. And AMD is hardly alone in this. It was Apple that originally developed OpenCL. That was back when they were still struggling to compete with Microsoft and were trying to convince developers to use open standards instead of DirectX. Once the iPhone became a dominant platform, they suddenly decided they didn't like open standards anymore and pushed everyone to use their proprietary APIs. Intel also used to be 100% dedicated to OpenCL, until they decided to create oneAPI instead. The reason NVIDIA has come to dominate the market is that they're the only ones who've been consistent over the long term. CUDA was introduced back in 2007 and they've stuck with it ever since. They also have consistently made good hardware that was well suited to HPC. Unlike, say, the Xeon Phi monstrosity Intel tried to dump on us. |
|
Maybe we can just consider it hypothetically. I'm not saying we have to implement a backend. It's also a good idea to wait, see if it really sticks. By the time it does, OpenCL might really start showing its wrinkles. The US recently banned exports of A100s and H100s to China, yet crippled their semiconductor production capabilities. Simple reasoning suggests they'll rise back up, with custom Chinese accelerators in ~5-10 years (as @DanielWicz mentioned). Will those fresh hardware support then-ancient standards like OpenGL and OpenCL? Right now I can maintain the Metal backend, maybe for 5 years, but not for 20. That's the same timeframe where we might see SYCL become established. In any case it's good to prepare for the future, anticipate how that might impact OpenMM. |
|
I'm certainly hoping for things to swing back toward standards again! |
|
Back to the original subject, it looks like the effect of adding the So for AMD, this probably isn't a useful optimization. If we can figure out why it produces a speedup in some cases and a slowdown in others, perhaps we can find a better approach that consistently makes it faster (or at least not slower). For Apple I think it could be useful. I believe it always produces a speedup on all computers we've tested? If so, we could perform the flushes on Apple GPUs only. |
Is limiting "internal" development to the Common and OpenCL platforms a realistic possibility?
Exactly. Hardware vendors made the mess and should, ideally, own the consequences as well. Meaning they should put the resources to support their hardware with their language inventions. But, to be fair, in this case, AMD cleaned up the mess a bit. The HIP work appears very good--judging only by the performance and ease of installation. The question is how much additional work has cost you to interact with them and how much more work it will take to fully incorporate their implementation into OpenMM. |
What if the community converged and made a SYCL backend as a plugin? HIP, Metal, and others would all be able to use the backend. |
|
There's a table in my computing center (CINES) that summarizes nicely all the frameworks |
|
@ex-rzr Thank you for the benchmarks of the three flush version. Would you also be able to run benchmarks with only 1 flush enabled? First try only before forcesKernel (Perm 1) and then only after clEnqueueReadBuffer (Perm 5). The extra pauses introduced by multiple flushes may be cancelling out the gains from a beneficial flush. |
|
MI100 8.0.0 before forcesKernel (Perm 1) after clEnqueueReadBuffer (Perm 5) MI210 8.0.0 before forcesKernel (Perm 1) after clEnqueueReadBuffer (Perm 5) V620 8.0.0 before forcesKernel (Perm 1) after clEnqueueReadBuffer (Perm 5) |
Perhaps try the optimization here? |
|
I used the changes in #3954 to profile on AMD. Here's a timeline for PME with the original code. There are gaps of 50-60 us after computeNonbonded and findBlocksWithInteractions. The timeline makes it look like there are also gaps after finishSpreadCharge and reciprocalConvolution, but there aren't really. Those are the FFTs. It only captures kernels launched through OpenCLContext, so it doesn't show the ones launched by VkFFT. Here's what it looks like with a There's no longer a significant gap after computeNonbonded, but instead we have a gap of a similar size immediately before it. So why is it faster? According to the profile, it isn't. It shows the time per step as basically unchanged at 1.1 ms. Which suggests that profiling is distorting the results and changing the thing I'm trying to measure. :( Here's the timeline for apoa1pme (original code). The same gaps are there as before. The one after findBlocksWithInteractions is about the same size as before, which makes it less significant in relation to the length of the kernels. The one after computeNonbonded has grown roughly in proportion to how long the kernel execution has grown, so it's still a similar fraction of the time. And here it is with No significant change to much of anything. |
|
@peastman This is really cool what you've put together! You may get a better idea of what's going on if you increase the precision of start time to string conversion to 8 digits. 6 digits says these started simultaneously: 8 digits shows 2-3 us apart: |
With |
Where did you find that? |
I mentioned it a while ago on the "future of osx GPU support" thread. That why initially, when I didn't know how the Mac OpenCL driver worked, I was worried. My hypothesis, it flushed after every command, causing severe CPU-side overhead. Switching to Metal would make the overhead preventable. Recently, you were there when I learned the Mac OpenCL driver does not do that. The driver does not flush after every command. That is, unless you enable profiling. The only way that's accomplishable, submit each command in its own command buffer. Time the beginning and end of the command buffer. Creating a new command buffer is by definition flushing the queue. This limitation is universal to GPU vendors, including AMD. this hyperlink -> Cmd + F + "Enabling profiling" |
|
That hypothesis were true, then adding the extra You're making assumptions about how the GPU works: that profiling is implemented entirely on the host side. In fact, many GPUs have hardware support for profiling and can automatically collect lots of metrics. For those GPUs, the profiling API is just a way of querying the data collected by the GPU. It need not have any impact on how the host dispatches work to it. |
- Implements perm 5 from openmm#3937 (comment) Co-Authored-By: Philip Turner <philipturner.AR@gmail.com>
|
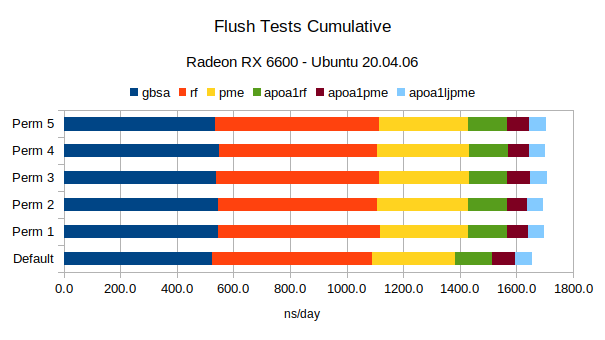
Linux flush tests results on RX 6600. Perm 3 and 5 provide the most consistent outcomes.
|
* Enable flush on Windows - Implements perm 5 from #3937 (comment) Co-Authored-By: Philip Turner <philipturner.AR@gmail.com> * Add brackets for clarification Co-authored-by: Philip Turner <philipturner.AR@gmail.com> * Make this optimization only apply to AMD GPUs * Switch to perm 1 - Flush before call to computeNonbonded since it works well on Windows and Linux * Update OpenCLNonbondedUtilities.cpp * Perm 4 is now significantly faster on Windows * Use isAMD * Fix indentation * Fix missed variable * Remove Mac check * Remove isAMD out of Mac code * Consistent (lack of) brackets style --------- Co-authored-by: Philip Turner <philipturner.AR@gmail.com>
|
We've made some substantial gains in OpenCL performance on AMD over the past 10 months! Thank you @philipturner for bringing great ideas and @peastman for your assistance implementing them.
|
|
That's excellent! Thanks for pushing on this. It's good to see we've made real progress. |
|
This is also great because I have an AMD 7900 XTX, just waiting until I need to run a larger simulation before I get the Linux box wired up. |
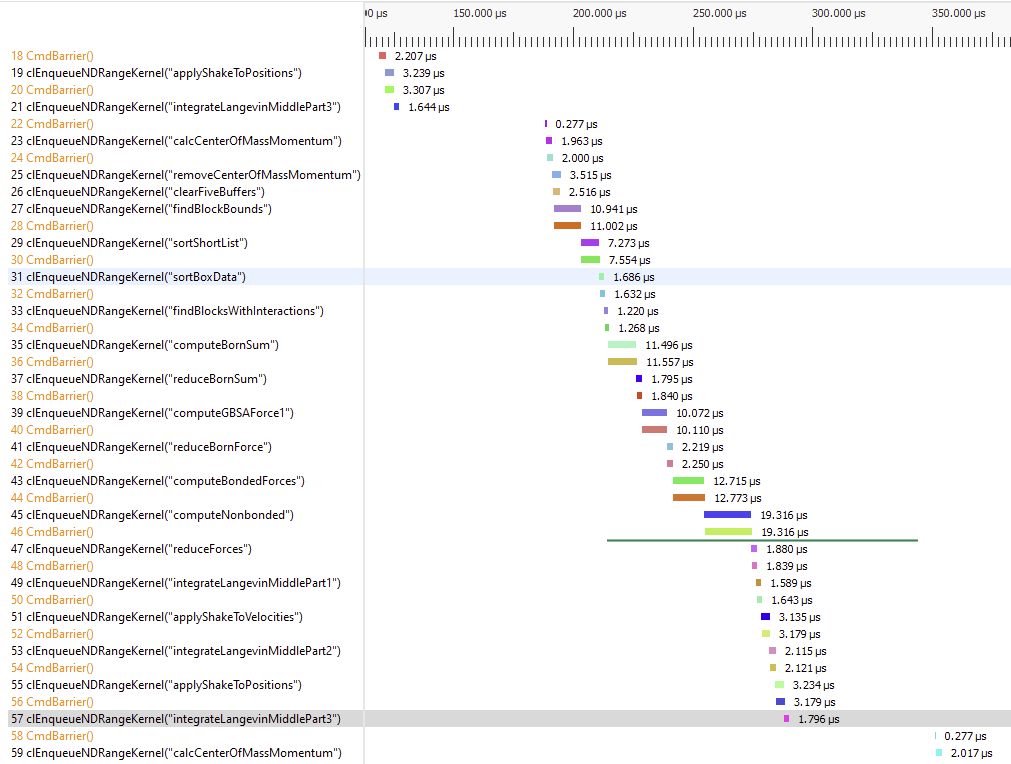

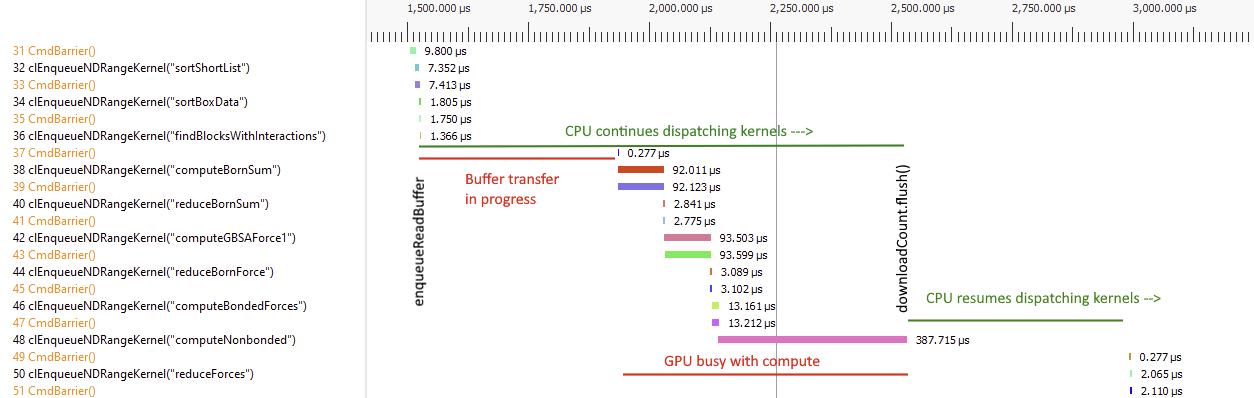
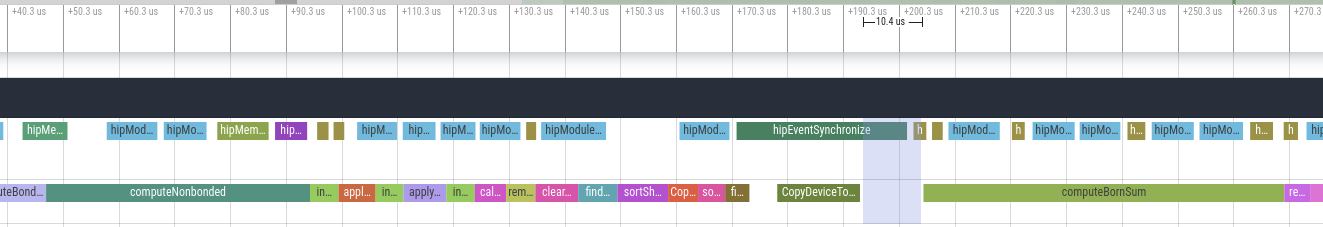


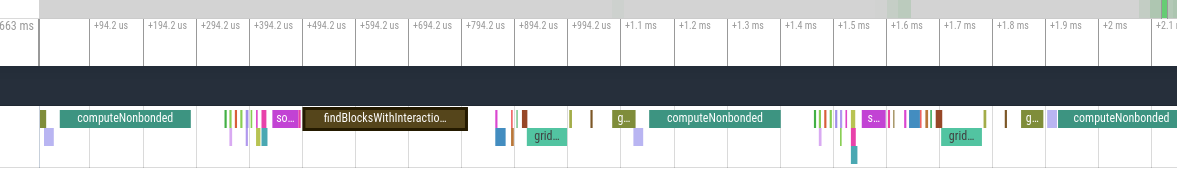





This is to continue the discussion that started in #3934. On AMD GPUs, the OpenCL platform is sometimes several times slower than the HIP platform. We're trying to figure out why. Much of the slowness seems to be due to large gaps between kernels where the GPU is sitting idle.
The text was updated successfully, but these errors were encountered: