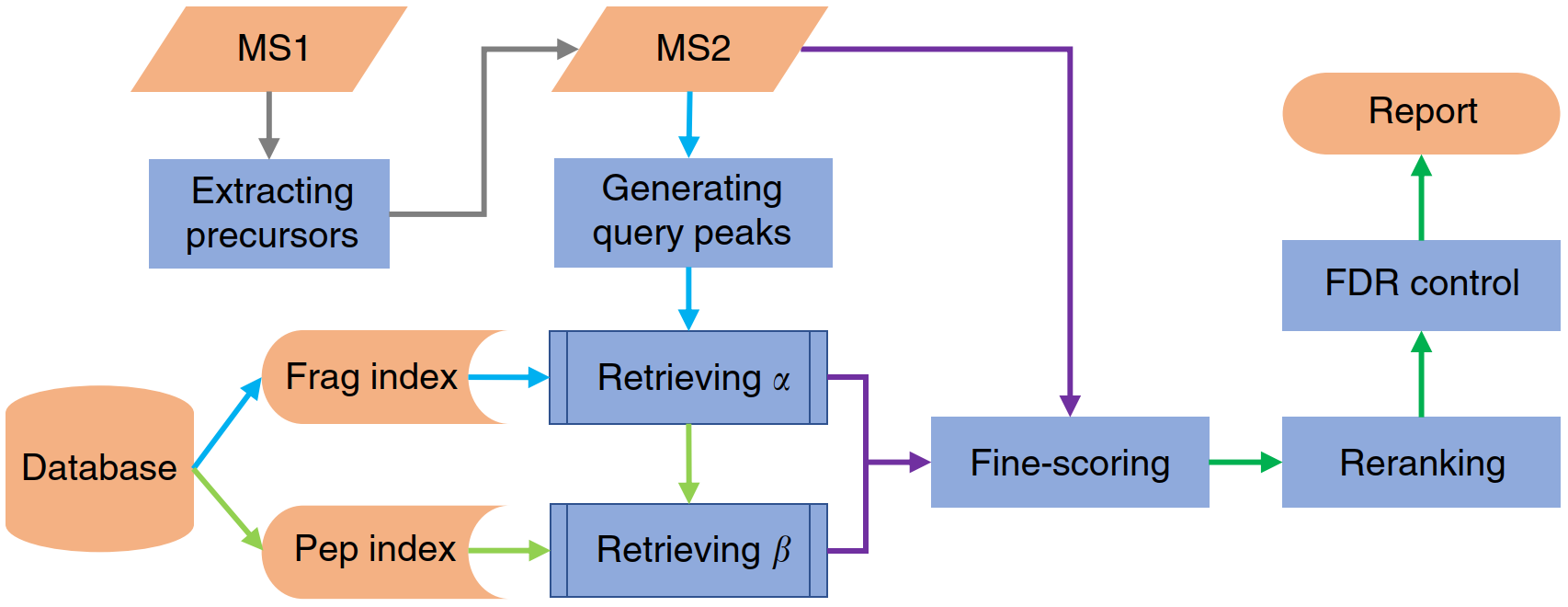
pLink® is a software dedicated for the analysis of chemically cross-linked proteins or protein complexes using mass spectrometry.
pLink 2 is developed as an upgrade of pLink 1. Compared with pLink 1, pLink 2 provides a graphical user interface, and is ~40 times faster with a newly designed index structure. There are also some improvements in the precision.
Our paper A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides has been accepted for publication by Nature Communications. Congratulations and please cite this paper if you used pLink 2.
A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Zhen-Lin Chen, Jia-Ming Meng, Yong Cao, Ji-Li Yin, Run-Qian Fang, Sheng-Bo Fan, Chao Liu, Wen-Feng Zeng, Yue-He Ding, Dan Tan, Long Wu, Wen-Jing Zhou, Hao Chi, Rui-Xiang Sun, Meng-Qiu Dong & Si-Min He. Nature Communications. July 30, 2019. [abstract]
Please download and read user_guide.pdf before download and use pLink 2.
pLink 2 is currently free to use. Download pLink 2.3.
If you have any questions about it, please contact pLink@ict.ac.cn.
Online discussion: https://github.com/pFindStudio/pLink2/issues, see github.pdf for usage.
- The expiration date of pLink 2.3.11 is set on Jan. 10, 2025.
- If RAW file read required, MSFileReader, 3.0 SP2 or a lower version, both 32 bit and 64 bit, need be installed first.
- If you have problem to run pLink 2 on Windows 7, please see this issue. Re-install MSFileReader using "Run as administrator".
- .NET framework 4.5 or a higher version is required.
- Java is also required for quantitation.
- Change the website to http://pfind.org from http://pfind.ict.ac.cn.
- Add the self-service registration website http://i.pfind.org.
- The license for version 2.3.10 is still valid for version 2.3.11. The same expiration date is Jan. 10, 2025.
- If you have problem to run pLink 2 on Windows 7, please see this issue. Re-install MSFileReader using "Run as administrator".
- Fixed a major bug in calculating E-value that causes software to crash, thank you Olexandr Dybkov and Liu Lab who reported.
- Changed the format of 'Precursor_Mass' and 'Peptide_Mass' in the report to decimal, thank you @IwanParf.
- Fixed several other bugs.
- All users are recommended to update to this version if required.
- A new license is required for this version, but we have extended the license validity to three years. This means you can use pLink 2.3.10 until 2025.1.10 without worrying about expiration.
- If you have problem to run pLink 2 on Windows 7, please see this issue. Re-install MSFileReader using "Run as administrator".
- Fixed a pParse bug, thank you lili.
- Fixed a fasta reading bug, thank you @vladtheimpalerSr.
- Updated the user guide, please read it before installation.
- All users using version 2.3.8 are recommended to update to this version.
- License for version 2.3.8 is still valid for this version.
- If you have problem to run pLink 2.3.9 on Windows 7, please see this issue.
- Happy New Year 2020!
- Fixed a quantitation bug on French Edition of Windows.
- Updated the configuration for cross-linker.
- Updated pLabel for MS-cleavable cross-linked spectra annotation.
- Updated the user guide, please read it before installation.
- A new license is required for this version, but we have extended the license validity to three years. This means you can use pLink 2.3.8 until 2023 without worrying about expiration.
- Fixed a bug when open pLabel.
- License for version 2.3.5 is still valid for this version.
- Congratulations! Our paper of pLink 2 is finally published, and please cite this paper if you used pLink 2.
- Fixed a bug when register the software.
- Fixed a bug when showing the symbol of β in pLabel.
- Disabled the SUMO identification flow temporarily, as we are optimizing and testing it. This feature will be available as soon as possible.
- Updated the lists of modifications and enzymes.
- Updated the GUI.
- License for version 2.3.5 is still valid for this version.
- Happy New Year 2019, and please see greetings from your pLink developers!
- Fixed a bug when infer proteins from peptides, thank you @zrpeak.
- Fixed a bug when the database is small.
- Fixed a bug when save a disulfide bond search task.
- Fixed a bug in modification tab of pConfig.
- Add a warning when combineSS.exe failed.
- Updated expiration time to January 1 2020.
- A new license is required since this version and will be valid until January 1 2020.
- Fixed a bug when searching super long peptides. Please note that the min length of peptides cannot be shorter than 4aa and the max length of peptides cannot be longer than 120aa.
- Fixed a bug when the unit of fragment tolerance is Da.
- Fixed a bug when activating software in Windows 7.
- Merged the SS flow and the SS_0 flow in disulfide bonds identification.
- Provided the global FDR estimation for intra-protein and inter-protein cross-links.
- Supported lower case amino acids in fasta file.
- Added a warning when the path of pf2 file is invalid.
- Improved GUI usability.
- Updated expiration time to August 1 2019.
- A new license is required since this version and will be valid until August 1 2019.
- Fixed a bug when labeling the -NH3/-H2O peaks in pLabel.
- Fixed a bug when drawing the FDR curve.
- Fixed a bug when running on Windows 10 Build 1803.
- Improved the spectra preprocessing algorithm.
- License since version 2.3.0 is still valid for this version.
- Fixed a bug when the m/z of a peak is negative.
- Fixed a bug when the result is empty.
- Fixed a bug when computing E-value with SS_0 linker.
- Fixed a bug when clicking HELP button in pLabel.
- Corrected the mono mass of SS_0 linker.
- Found solution to pLabel's annotation problem on non-Chinese Edition of Windows.
- License since version 2.3.0 is still valid for this version.
- Fixed a bug on French Edition of Windows.
- License for version 2.3.0 is still valid for this version.
- Fixed the decimal point bug on French Edition of Windows.
- Fixed a bug on some Windows 10 Pro.
- Fixed bugs when configuring Enzymes and Quantifications.
- Fixed pLabel annotation problem with MGF extracted by PD.
- Disabled the requirement of administrative privileges.
- Improved software activation methods.
- Improved robustness when searching against *.fasta file containing non-alphabet characters.
- Improved GUI usability.
- Added validity check when configuring meta data.
- Extended the max missed cleavages to 5.
- Extended quantification support to BS3 labeling.
- Changed the default variable modification when linker SS used.
- Removed pBuild folder in results.
- Updated the user guide.
- First public beta version.