-
Notifications
You must be signed in to change notification settings - Fork 3
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
1 changed file
with
51 additions
and
1 deletion.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,4 +1,54 @@ | ||
| # Data analysis | ||
|
|
||
| TBD | ||
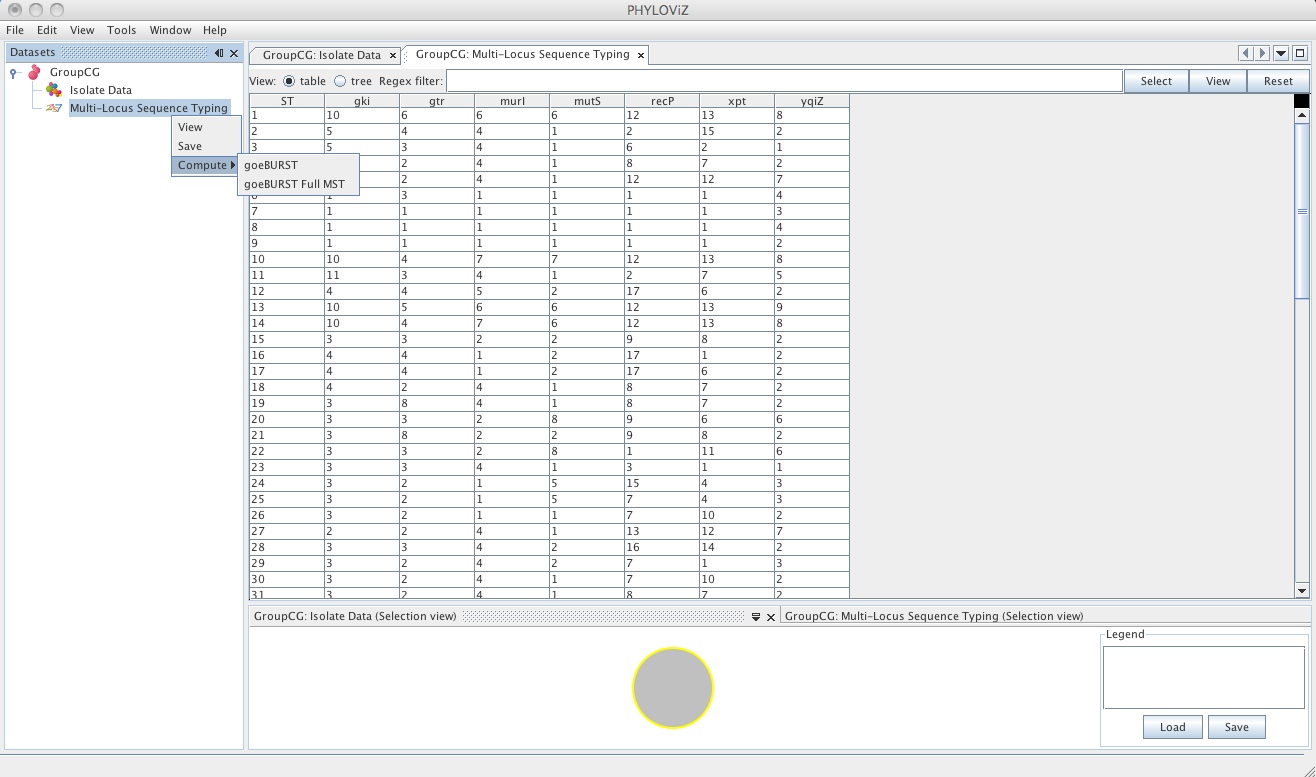
| In the current version of PHYLOViZ, you can analyze your data using the goeBURST algorithm and an extension of the goeBURST rules to draw a full Minimum Spanning Tree (MST). Press the _Right Mouse Button_ on the _Typing Data_ (now named with the method) and choose compute to access the available analysis algorithms. | ||
|
|
||
|  | ||
|
|
||
| ## goeBURST algorithm | ||
|
|
||
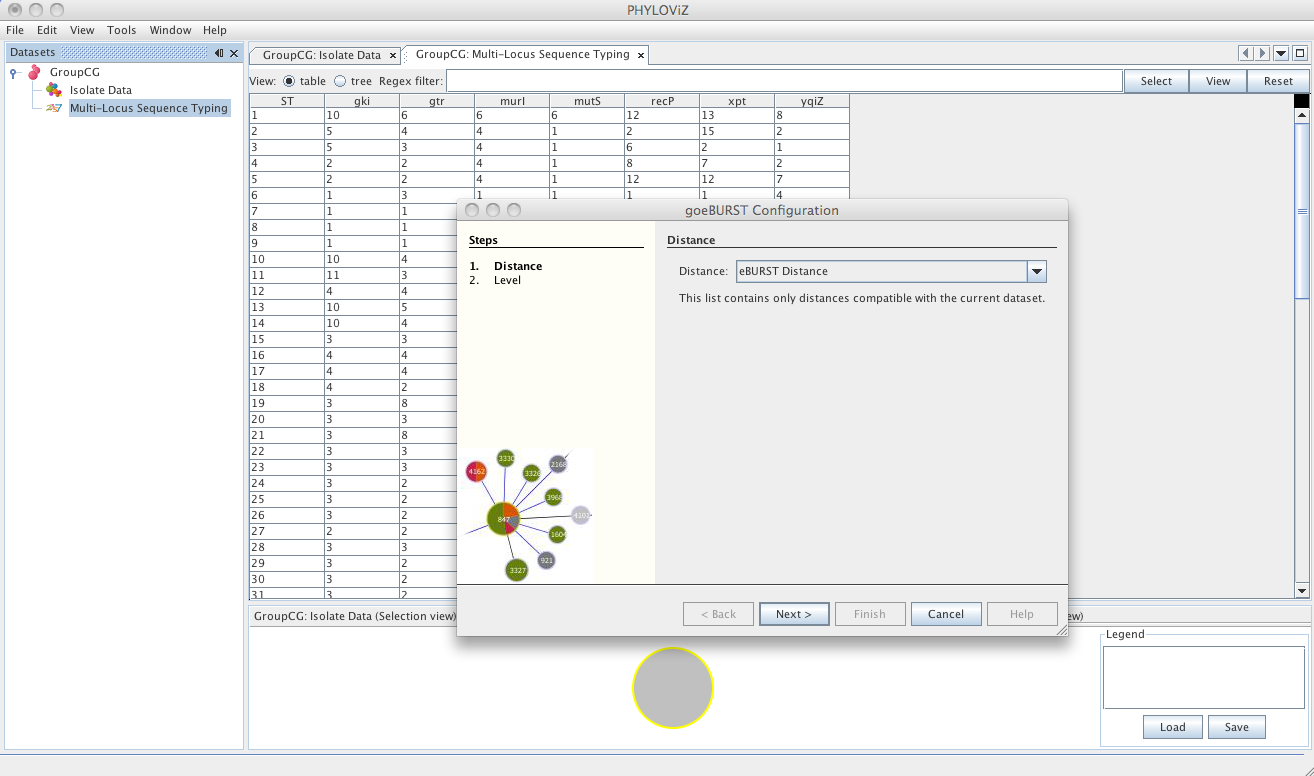
| Selecting the goeBURST algorithms opens the dialog for the goeBURST algorithm. The algorithm was originally described in the article [Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach](http://www.biomedcentral.com/1471-2105/10/152). The first step is choosing the _Distance_ to be used. Currently eBURST Distance is the only one available, but others could be implemented. The eBURST distances follows the tiebreak rules discussed in the article. | ||
|
|
||
|  | ||
|
|
||
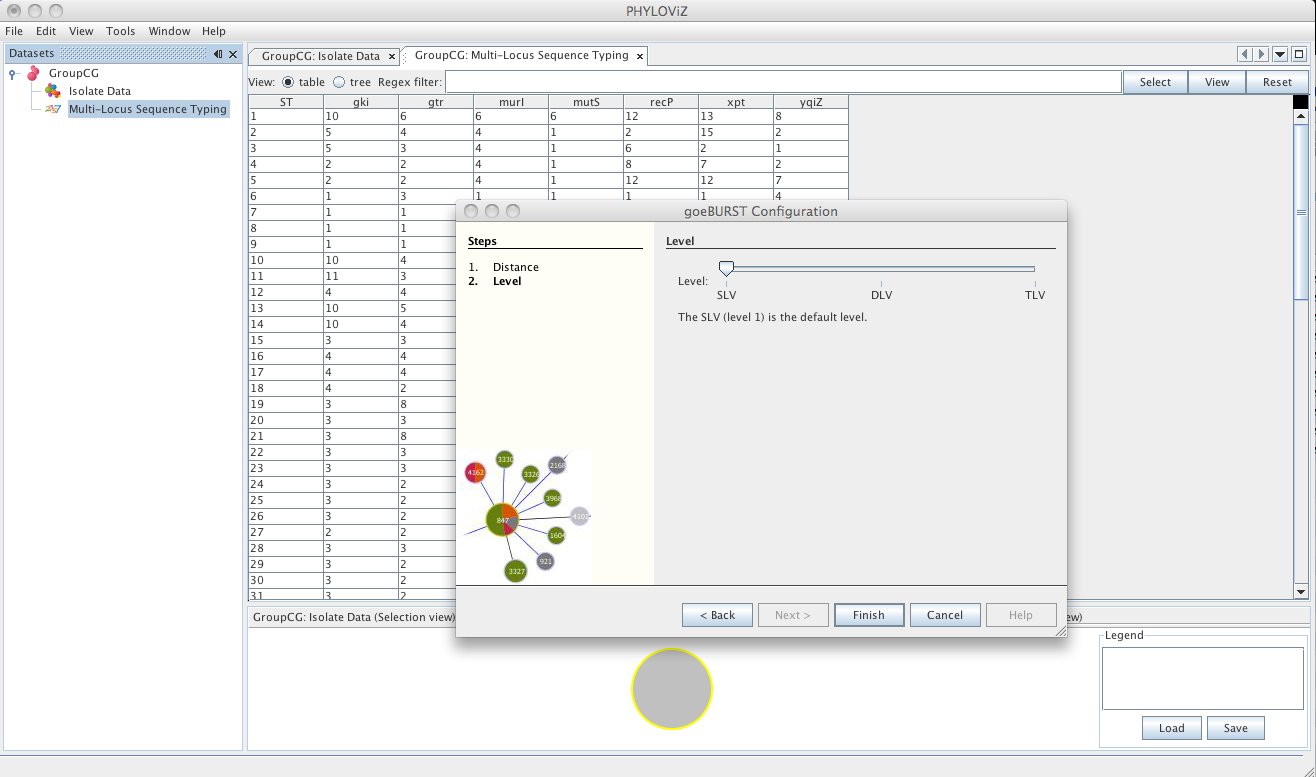
| The second step is the choice of the level to which clonal complexes will be formed. The usual default for MLST analysis is SLV Level. Choosing DLV or TLV level will take longer calculation times, but could provide some insight to the relationships between clonal complexes formed at SLV and DLV level respectively. | ||
|
|
||
|  | ||
|
|
||
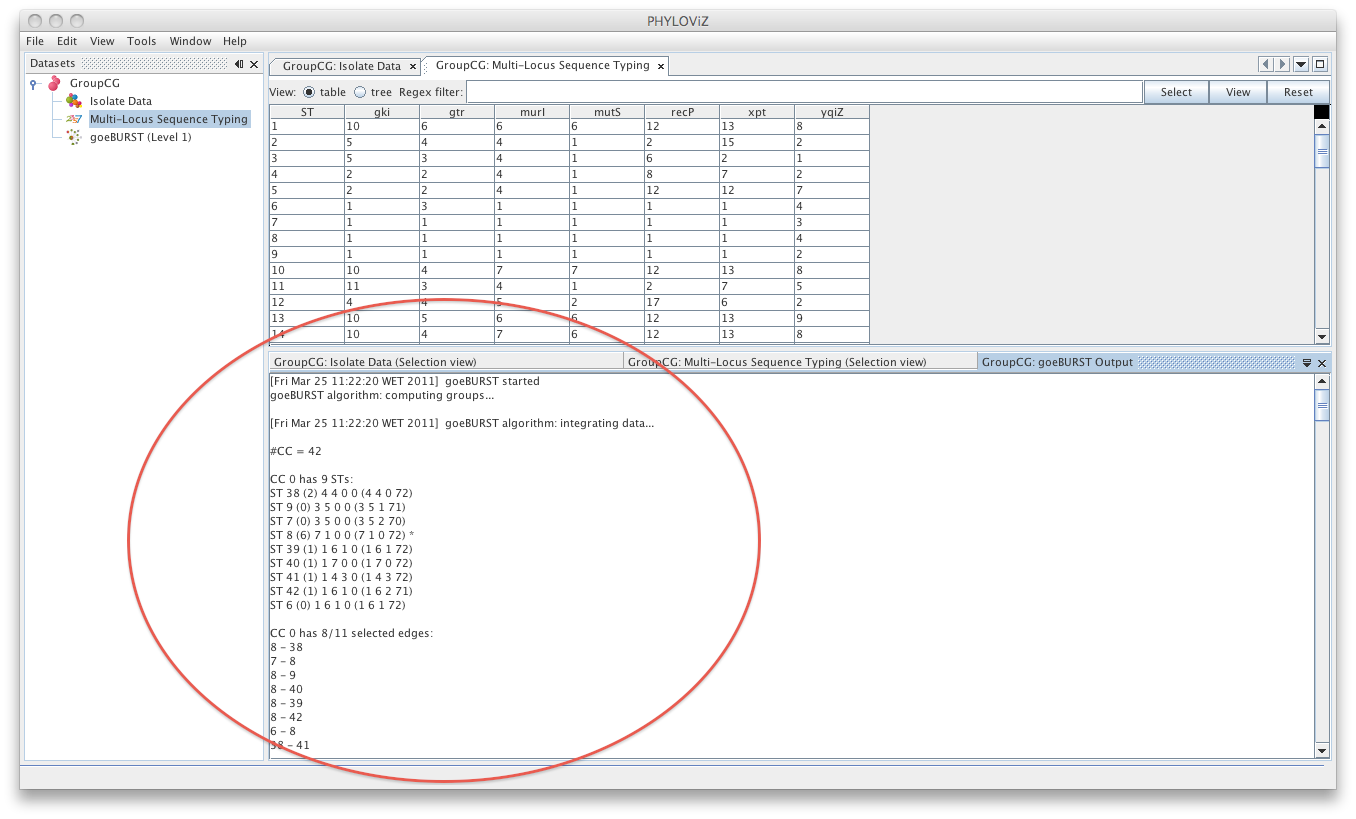
| A goeBURST _Output_ tab will appear and display the goeBURST algorithm results. It will contain information about the Clonal Complexes (CCs), namely the Sequence Types that compose them and what edges (the links between STs) were drawn in each CC. | ||
|
|
||
|  | ||
|
|
||
| Double clicking on the goeBURST item that is now on the Dataset tree menu will show the display. The clonal complexes will be arbitrarily numbered starting from 0 (for the CC with most STs) and contains all the data relevant to the goeBURST analysis (STs in each group and the drawn SLVs edges). The following screenshot summarizes the output for a single clonal complex with the test dataset used. | ||
|
|
||
| 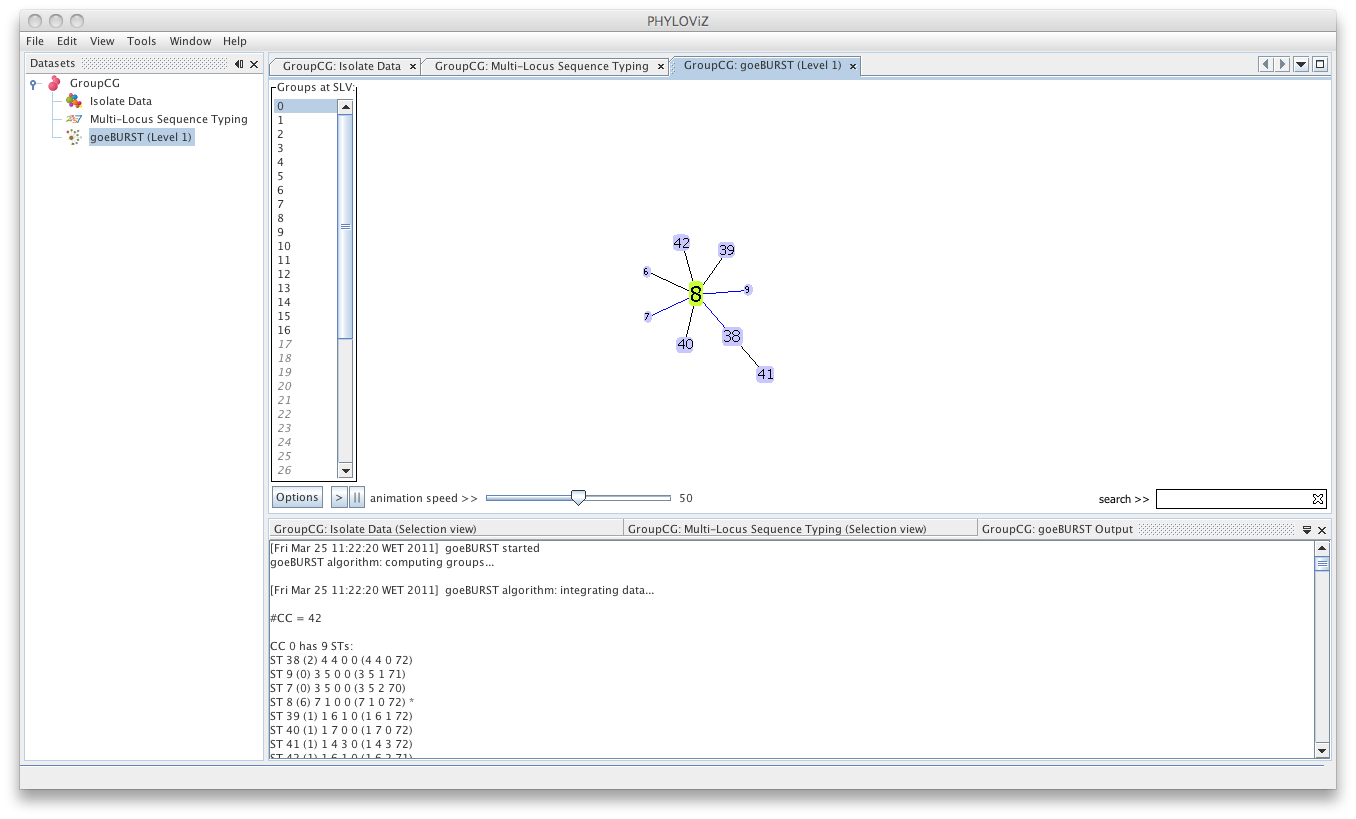 | ||
|
|
||
| Multiple groups can be displayed simultaneously by selecting them, using the CTRL /CMD and/or SHIFT keys. | ||
|
|
||
| 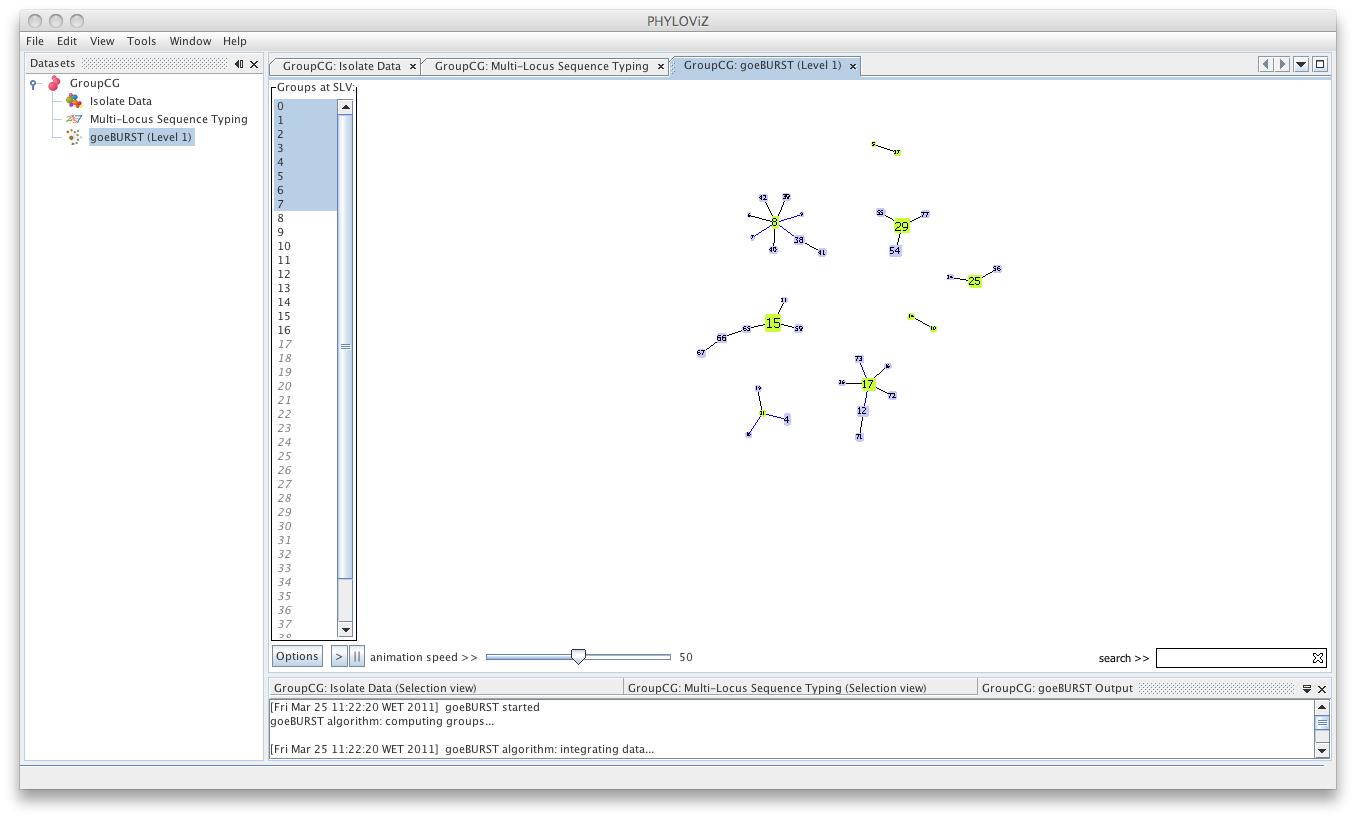 | ||
|
|
||
| ## goeBURST Full MST algorithm | ||
|
|
||
| Using an extension of the goeBURST rules up to \\(n\\)LV level (where \\(n\\) equals to the number of loci your dataset uses), a Minimum Spanning Tree-like structure can be drawn. Select _goeBURST Full MST_ in the _Compute_ options to draw it. Contrary to the standard goeBURST, the link statistics are not presented. After computation, double click on the _goeBURST Full MST_ that appears under the dataset heading to visualize the result. | ||
|
|
||
| 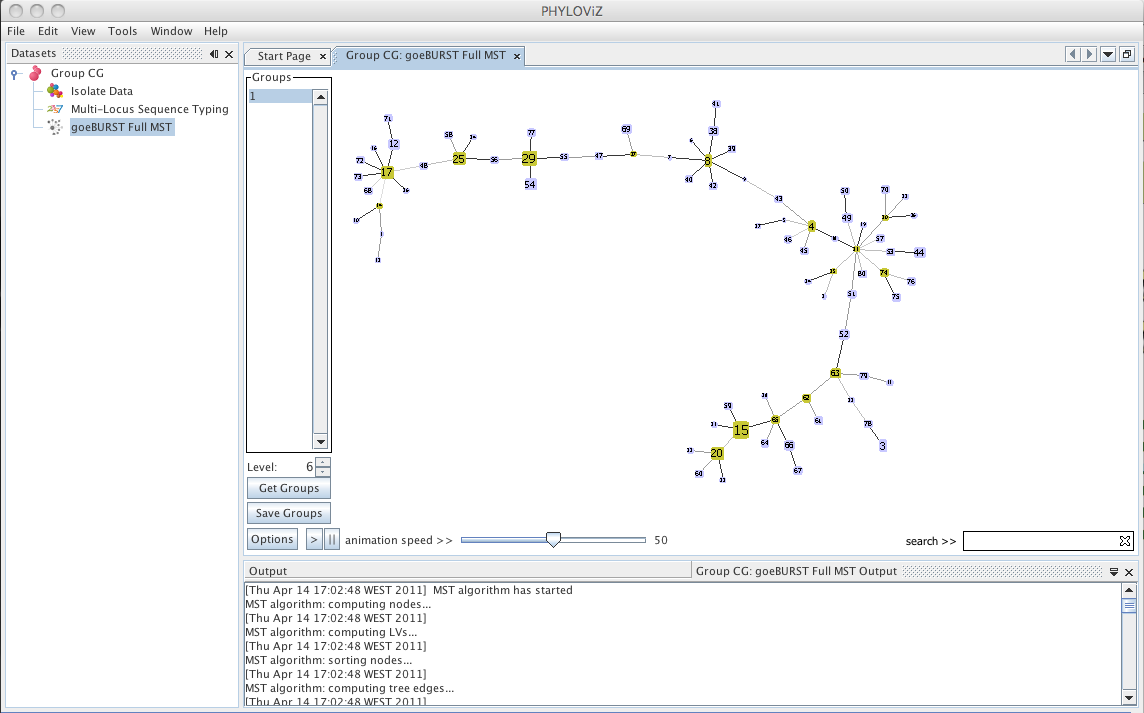 | ||
|
|
||
| New options appear on the display: The _Level_ selector and two new buttons _Get Groups_ and _Save Groups_. The _Level_ represents the _Locus Variant_ level and allows the removal of all the links greater than the number represented. The user can use the up and down arrows or directly edit the number by clicking on it. The _Get Groups_ button allows separate the display of groups that are not connected at the level chosen in order to simplify the analysis of larger datasets. This will generate a display very similar to that of goeBURST, but at a higher link level. The Save Groups creates an extra column in the isolate data with the title label _goeBURST MST[\\(x\\)]_ with \\(x\\) being equal to the level used to create the groups. | ||
|
|
||
| Decreasing the _Level_ selector, allows the user to see how clonal complexes would relate to each other at a certain level. Level 1, 2 and 3 are equivalent to calculating goeBURST at those levels (SLV,DLV and TLV respectively). The following images shows what happens to the dataset when you decrease the level. Level 4 is not displayed since no new groups are formed at that level. | ||
|
|
||
| 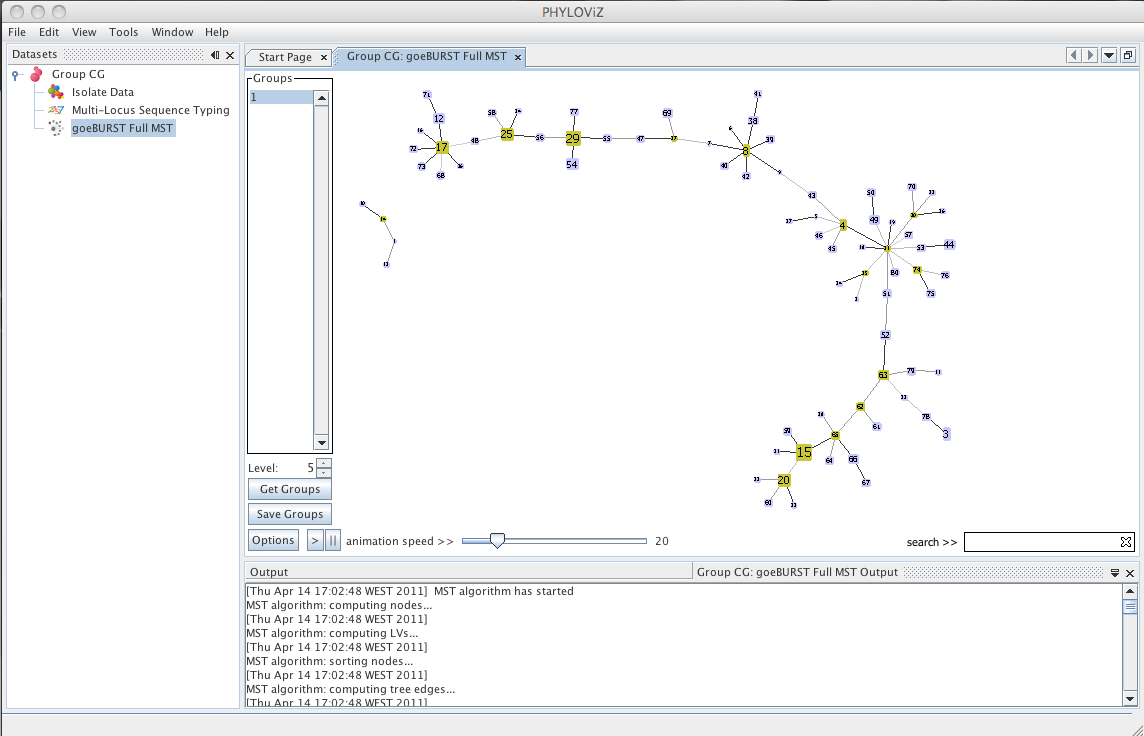 | ||
|
|
||
| At level 5 only two groups are formed in the sample dataset. | ||
|
|
||
| 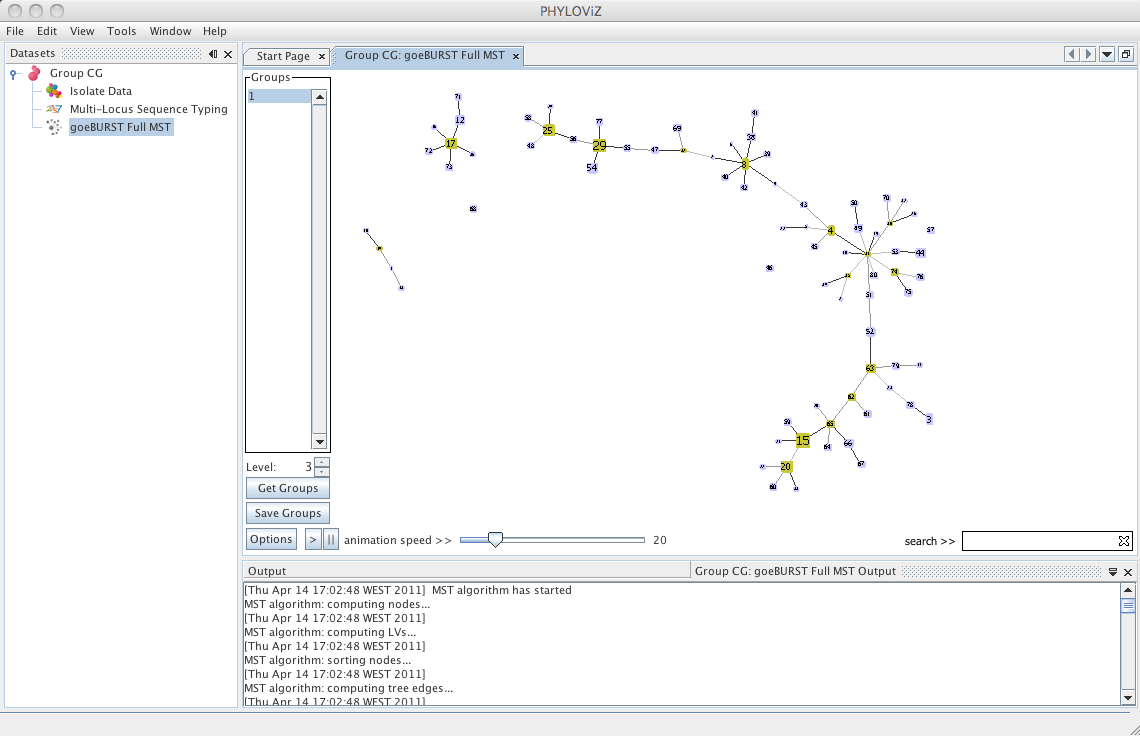 | ||
|
|
||
| At level 3 (TLV level) some singletons appear. Level 4 is not shown since no changes were observed in the graph. This means that there are no two STs in the dataset that differ in 4 of the loci of their profiles. | ||
|
|
||
| 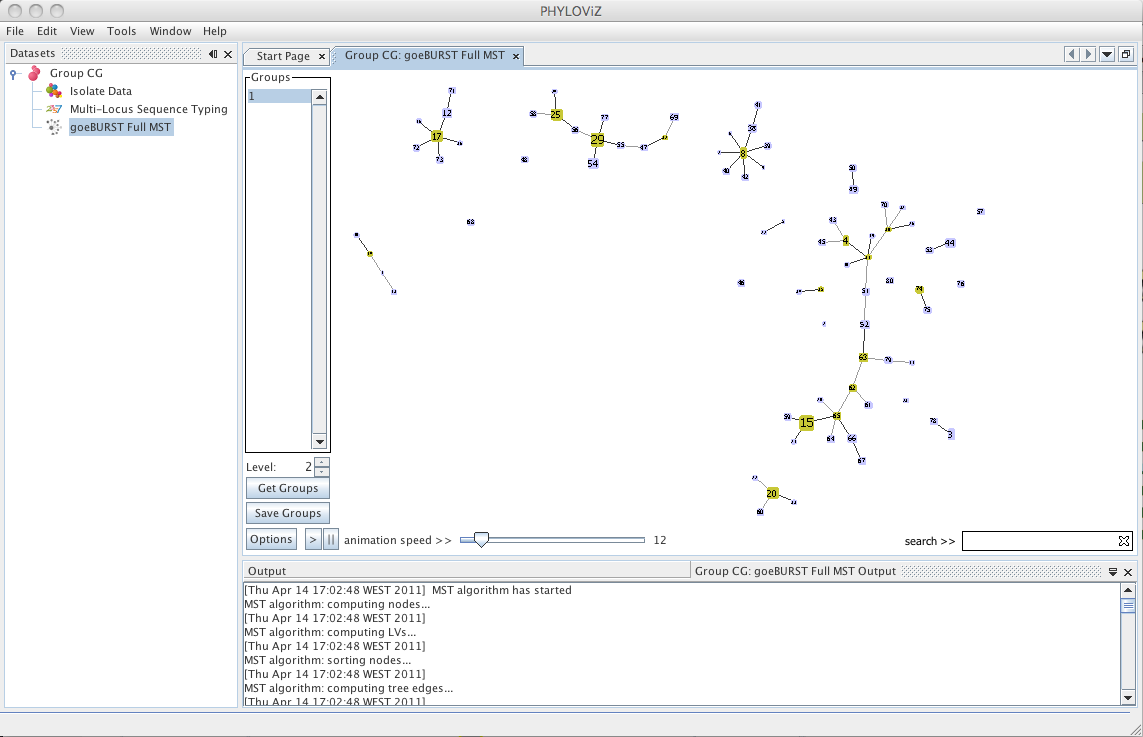 | ||
|
|
||
| At level 2 , 6 groups appear with 4 or more STs each. | ||
|
|
||
| 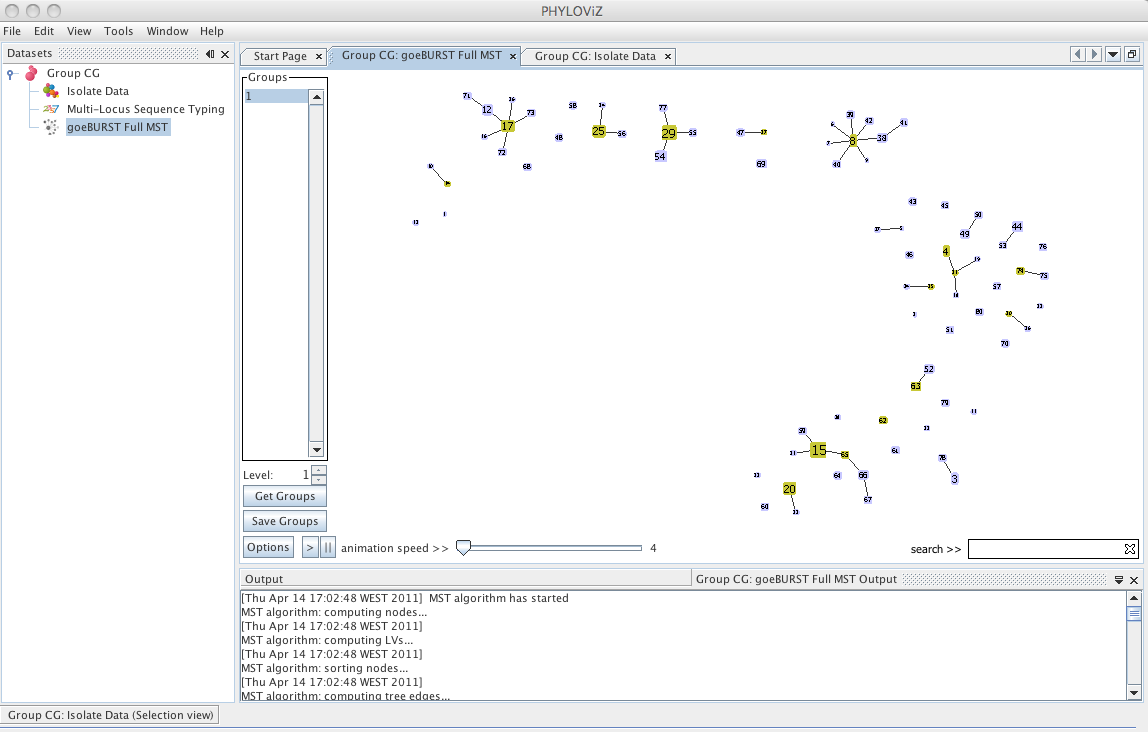 | ||
|
|
||
| And finally at level 1, the equivalent of the most commonly used Clonal Complex definition by goeBURST, 17 groups with 2 or more STs are formed and there are 25 singletons on the dataset. | ||
|
|