Subscribe for notifications about new releases 🚀
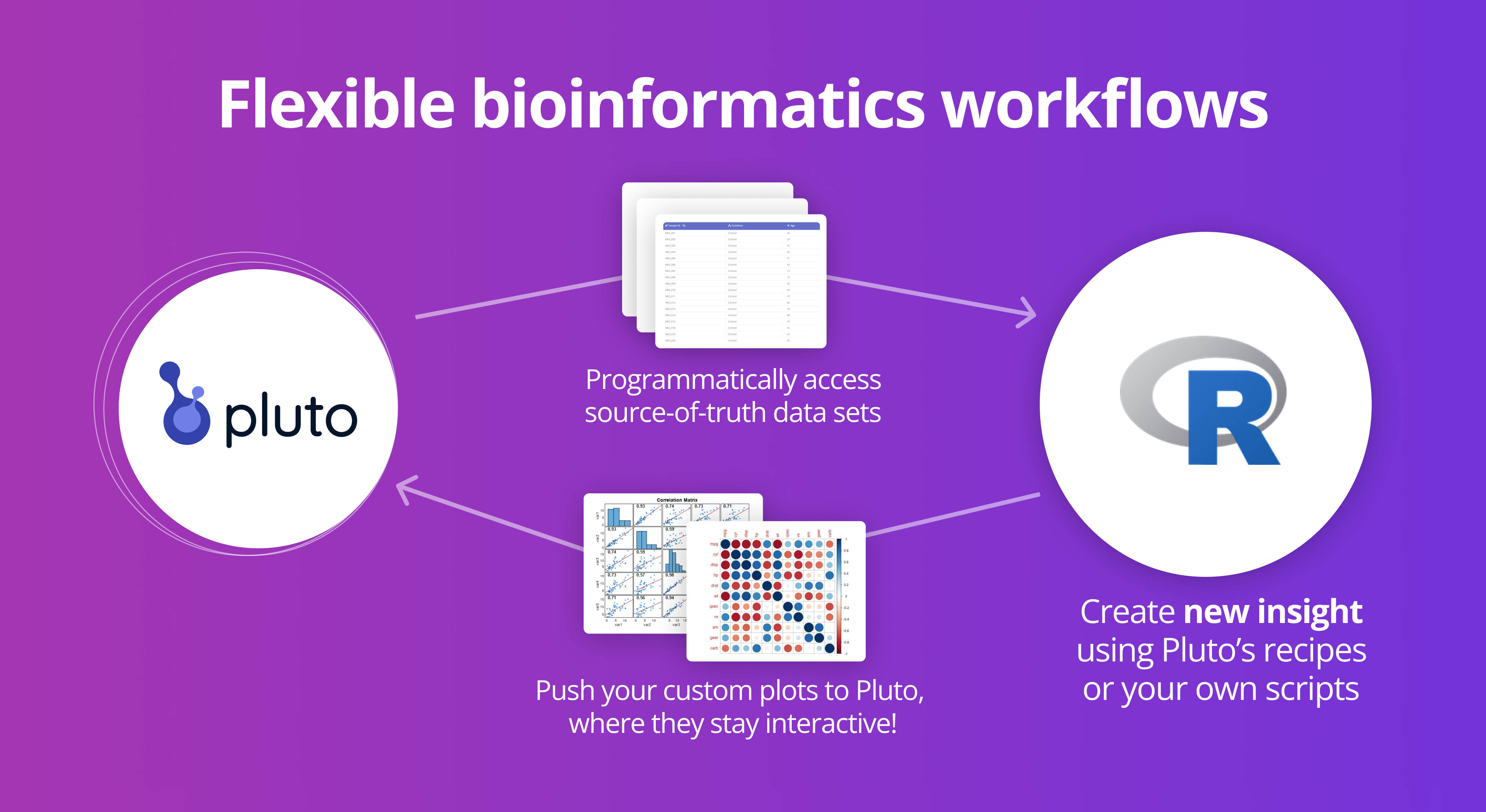
pluto is the official R package for interacting with Pluto, the biological discovery platform. With a few lines of code, you can read data and results directly into your R scripts, and push custom plots back to Pluto where they remain interactive and collaborative.
Not using Pluto yet? We'd be happy to show you the platform in action - schedule a personalized demo.
Install the latest version from Github:
# Install devtools if you don't already have it
install.packages("devtools")
# Install the pluto R package
remotes::install_github("pluto-biosciences/pluto-sdk-r")
Once installed, you can load the Pluto R package into your scripts with library(pluto).
A typical, minimal workflow using pluto involves:
- Authenticate with Pluto API token (see
vignette("authentication")) - Fetch data from one or more experiments
- Perform some custom analysis (see
vignette("rnaseq_recipes")for inspiration ✨) - Push your results back into the Pluto canvas
There are also numerous ways to use pluto at a larger scale, for example with multiple data sets or integrating with your in-house tools. A few examples include:
- Fetching all RNA-seq experiments in a project and performing a meta-analysis of their differential expression results
- Fetching four different NGS experiments (e.g. RNA-seq, ChIP-seq, CUT&RUN, and ATAC-seq) and overlapping differential regions to identify regions where both expression and binding is altered
With Pluto, your cross-functional team has the power to collaborate easily in one place regardless of coding ability. The same applies to your vendors as well!
Instead of having your outsourced bioinformatics vendor send you results by email or in folders where critical findings are easily lost, you can request that they deliver data and results directly back into your Pluto lab space. No training necessary on your part - Pluto handles all onboarding and training for bioinformatics CROs collaborating with clients in Pluto as a part of the service. So you can sit back and watch the results come to you. Chat with our Customer Experience team to learn more.
We're here to help! Feel free to reach out to support@pluto.bio or email your Pluto customer representative directly.
When running tests and developing the pluto R package, you will need to decrypt the encrypted .Renviron file in this repo.
Decrypt the encrypted .Renviron file
You will need to decrypt the .Renviron file the first time you start working in this repo, and any time other developers have made changes. It's good practice to check regularly for updates.
openssl enc -d -aes256 -base64 -in .Renviron.encrypted -out .Renviron
If you make modifications to the .Renviron file (e.g. adding new experiments for testing, or other variables), encrypt your file and check it in
Encrypt the modified .Renviron file
openssl enc -aes256 -base64 -in .Renviron -out .Renviron.encrypted
Check in the updated .Renviron.encrypted when opening a PR so that other developers have the latest changes.
To run tests: devtools::test()
Note that most tests in the test suite are integration tests, so they require a specific API token and uuids present in the .Renviron file.
To build pkgdown site: pkgdown::build_site()