-
Notifications
You must be signed in to change notification settings - Fork 3
Comparison mode
Due to different networks are constructed from different data, it is difficult to compare these networks between them, because, for example, in case of protein nodes, one protein node in network 1 can be composed by protein A, protein B and protein C because they share all their peptides, and in network 2, due to the detection of a peptide unique to protein C, we will have protein A and protein B in one single protein node and protein C in another protein node.
These situations are difficult to manage when we want to know which proteins or peptides are in common or different between two networks, and ProteinClusterQuant identify all these nodes as the same, in order to get a rough estimation of the comparison. ProteinClusterQuant comparison mode perform a comparison of different networks created by the analysis mode in order to detect which are the peptide nodes or protein nodes that are in common or are different between these networks. It basically construct a Venn diagram between the networks reporting the overlapping nodes and unique nodes.
A template for this input parameter file can be downloaded here.
# Note: lines starting with '#' are ignored
######################################################
# Comparison type: Compare peptide nodes ('PEPTIDE_NODES') or protein nodes ('PROTEIN_NODES')
######################################################
comparison_type PROTEIN_NODES
######################################################
# FDR threshold: only consider the entries with a FDR value greater than the threshold
######################################################
threshold 0.05
######################################################
# INPUT FILES TO COMPARE
# Each file contains two columns, separated by a TAB.
# first column correspond with the name of the dataset
# second column correspond with the full path to the file corresponding to the FDR output file of a PCQ analysis, which typically is named as: 'prefix_FDR_sufix.txt' where prefix and sufix correspond to the input analysis parameters: 'output_prefix' and 'output_sufix'
######################################################
dataset1 /path_to_significance_files/prefix1_FDR_suffix1.txt
dataset2 /path_to_significance_files/prefix2_FDR_suffix2.txt
dataset3 /path_to_significance_files/prefix2_FDR_suffix2.txt
Where:
- comparison_type indicates if the comparison is going to be performed at protein_node or peptide_node level
- threshold indicates which portion of the datasets are taken into account in the comparison by defining a minimum FDR threshold.
-
dataset1, dataset2 and dataset3are arbitrary names assigned to each ones of the datasets to compare. They are separated by a TAB from the path where the output FDR file is located. Note that each dataset is indicated in a different line and that 2 or 3 datasets are allowed to be compared.
Example of output of a comparison between two networks at protein node level:
###############
Comparison made on: Tue Sep 13 17:45:56 PDT 2016
Datasets compared:
RW C:\Users\Salva\Desktop\Dropbox\papers\SCRIPPS\Protein Cluster Quant\second submission\Suplemental Networks\SuppNetwork8_FibulinRW_OLD_DB_3PSMsPerPeptide_2016-09-13-15-07-38\FibulinRW_OLD_DB_FDR_3PSMsPerPeptide.txt
WT C:\Users\Salva\Desktop\Dropbox\papers\SCRIPPS\Protein Cluster Quant\second submission\Suplemental Networks\SuppNetwork9_FibulinWT_OLD_DB_3PSMsPerPeptide_2016-09-13-16-45-25\FibulinWT_OLD_DB_FDR_3PSMsPerPeptide.txt
lacZ C:\Users\Salva\Desktop\Dropbox\papers\SCRIPPS\Protein Cluster Quant\second submission\Suplemental Networks\SuppNetwork10_FibulinLacZ_OLD_DB_3PSMsPerPeptide_2016-09-13-17-17-11\FibulinLacZ_OLD_DB_FDR_3PSMsPerPeptide.txt
Venn diagram URL (copy and paste the following URL in a browser to get the Venn Diagram):
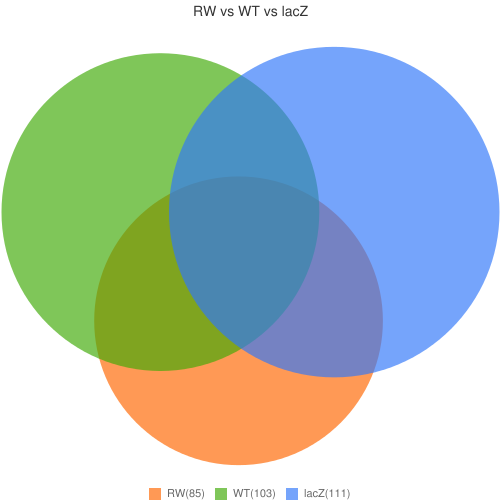
http://chart.apis.google.com/chart?chs=500x500&chd=t:85,103,111,37,36,33,26&cht=v&chdl=RW(85)|WT(103)|lacZ(111)&chtt=RW vs WT vs lacZ&chds=0,111&chdlp=b&chma=|10,10
1 -> RW = 85 (39.2% of union)
2 -> WT = 103 (47.5% of union)
3 -> lacZ = 111 (51.2% of union)
Union=217 (100%)
Overlap (1,2) = 37 (24.5% of Union (1,2))
Union (1,2) = 151 (69.6% of Total Union)
Overlap (1,3) = 36 (22.6% of Union (1,3))
Union (1,3) = 159 (73.3% of Total Union)
Overlap (2,3) = 33 (18.2% of Union (2,3))
Union (2,3) = 181 (83.4% of Total Union)
Overlap (1,2,3) = 26 (12% of union)
Just in 1 = 38 (17.5% of union) (55.3% overlapped)
Just in 2 = 58 (26.7% of union) (42.7% overlapped)
Just in 3 = 66 (30.4% of union) (38.7% overlapped)
The following lists correspond to the different overlappings between the lists, printing the PROTEIN_NODES name and the FDR value:
Intersection (26):
1 [ASNS 1.49902952973 FDR=1.13667556878E-6 Var=0.028473747332883083 #psms=6]
2 [HMGCS1 1.42141484575 FDR=2.5399628065E-6 Var=0.024902904950665744 #psms=20]
3 [VLDLR 0.915525369242 FDR=0.0196174897496 Var=0.024112519393382843 #psms=21]
4 [FADS2 2.22405279889 FDR=1.68101896039E-12 Var=0.04424805751985458 #psms=6]
5 [DST,THBS1,THBS2 1.26966472222 FDR=7.50112457161E-5 Var=0.025320653756107533 #psms=13]
6 [SMAD1,SMAD2,SMAD3,SMAD5,SMAD9 1.41570255199 FDR=1.34133634962E-4 Var=0.04427339030925525 #psms=4]
7 [FN1 1.22401485592 FDR=1.50588967253E-4 Var=0.02526431163855654 #psms=16]
8 [MFGE8 -1.05339070357 FDR=1.51994493815E-4 Var=0.021359464793734212 #psms=60]
9 [CBS 1.31220456145 FDR=2.76860547988E-4 Var=0.03798316832228635 #psms=6]
10 [GJA1 1.15276544598 FDR=5.20437298797E-4 Var=0.024666236061038762 #psms=12]
11 [COL11A1 1.05892733664 FDR=0.00297426076227 Var=0.025231834671429643 #psms=13]
12 [BCAT1 1.02362037365 FDR=0.00407878812159 Var=0.023293889600406355 #psms=39]
13 [GYS1 0.979472283557 FDR=0.00674857618242 Var=0.021779384524497774 #psms=33]
14 [GALNT1 1.00326848501 FDR=0.00713868056622 Var=0.025107947853406597 #psms=16]
15 [FTH1 -0.774886694325 FDR=0.0205488807321 Var=0.023449106752135115 #psms=29]
16 [P4HA2 0.831895627217 FDR=0.0459147227959 Var=0.022606449984463615 #psms=53]
17 [HSPG2 0.822185326135 FDR=0.0425163241433 Var=0.02037588604083698 #psms=85]
18 [ORC5,PLOD2 0.908568033401 FDR=0.0195531339311 Var=0.022716518697170472 #psms=33]
19 [ITGA5 0.908037081208 FDR=0.0191500069876 Var=0.02272151114781399 #psms=23]
20 [ERO1A 0.908665848157 FDR=0.0161862170785 Var=0.020844955090628194 #psms=96]
21 [SPARC 0.950883553024 FDR=0.0111899842286 Var=0.022802623017007195 #psms=22]
22 [PSAT1 1.24102075708 FDR=7.4211857087E-5 Var=0.022925350951431393 #psms=39]
23 [LOXL2 1.27082317853 FDR=7.22712288277E-5 Var=0.025910956375769206 #psms=16]
24 [MOV10 1.32453735232 FDR=4.64288832852E-5 Var=0.028078837418580277 #psms=11]
25 [CA9 1.26033019611 FDR=4.56831670018E-5 Var=0.022268383197575743 #psms=35]
26 [FTL -1.24215851511 FDR=5.05826927554E-6 Var=0.023376404983415942 #psms=22]
Unique to: RW (38)
1 [MIF -1.39991724426 FDR=1.15419136953E-5 Var=0.04156702461964517 #psms=16]
2 [BRD3 -1.28708179289 FDR=7.4609622438E-5 Var=0.040348692471837505 #psms=7]
3 [FUT8 -0.874255864619 FDR=0.0106682080581 Var=0.029144273740259904 #psms=3]
4 [COG7 1.07566957677 FDR=0.0154886692694 Var=0.04280344612531006 #psms=3]
5 [MMP15 1.05167507833 FDR=0.0174664141215 Var=0.041319993863463085 #psms=3]
6 [GPNMB -0.876041057573 FDR=0.0176133612874 Var=0.034813784650142184 #psms=12]
7 [EFEMP2 1.05043090361 FDR=0.0176882554825 Var=0.04094249780651749 #psms=3]
8 [FIS1 -1.12029423372 FDR=0.0299608115494 Var=0.08582731016174798 #psms=3]
9 [NT5C -0.836414598203 FDR=0.0347546818332 Var=0.03910383836366322 #psms=7]
10 [NDUFAF1 -0.834554596928 FDR=0.0397832342827 Var=0.04091606947523016 #psms=5]
11 [FAM21A,FAM21B,FAM21C -1.15200309345 FDR=0.0459317236962 Var=0.105166181451179 #psms=6]
12 [NUDCD3 -0.843891090087 FDR=0.0486112697482 Var=0.045947108406931794 #psms=3]
13 [NDRG4 -0.8769112416 FDR=0.0295079408825 Var=0.04275519874493582 #psms=6]
14 [PRKDC,VPS26A -1.59946207042 FDR=1.52264904296E-4 Var=0.08515785286042672 #psms=9]
15 [GNB1,GNB2,GNB3,GNB4 -1.18445082279 FDR=3.84039469998E-4 Var=0.040533537448000846 #psms=8]
16 [SMPD4 -1.18702400593 FDR=6.17132040221E-4 Var=0.04436333343366013 #psms=3]
17 [MRE11A -1.13239000806 FDR=7.37421465342E-4 Var=0.03922927836657316 #psms=3]
18 [CBLB,DKFZp779A0729 1.28443236353 FDR=9.56687948391E-4 Var=0.043936364678503335 #psms=4]
19 [ENPP2 1.09589535892 FDR=0.0103488176527 Var=0.04040939267152964 #psms=6]
20 [CTSD -0.939198709521 FDR=0.0103442469509 Var=0.03736624694595215 #psms=19]
21 [SIRPA,SIRPB1 -1.251538767 FDR=0.0101916096821 Var=0.08598514191219057 #psms=9]
22 [PSPH 1.09718741229 FDR=0.0100168892503 Var=0.04064353667641737 #psms=4]
23 [JAK1 -1.01551459701 FDR=0.00604171143555 Var=0.04183440192534903 #psms=3]
24 [SUPV3L1 -0.936470138905 FDR=0.00434740305413 Var=0.028998920002867374 #psms=8]
25 [NIT1 -1.65953438176 FDR=6.47338891241E-8 Var=0.042735512374216515 #psms=7]
26 [GPT2 1.7904557538 FDR=4.63065928269E-8 Var=0.041316152895064184 #psms=3]
27 [UBE4A 1.99486165873 FDR=2.98105232807E-10 Var=0.04147091556788849 #psms=11]
28 [UBL7 -2.85048564521 FDR=0.0 Var=0.0774801670969568 #psms=5]
29 [THUMPD3 -3.44509001378 FDR=0.0 Var=0.042739289477418854 #psms=4]
30 [LMF2 -3.83650126772 FDR=0.0 Var=0.08637510164802702 #psms=3]
31 [MYH10,MYH11,MYH14,MYH9 -5.05889368905 FDR=0.0 Var=0.0850207706885046 #psms=7]
32 [STAU1 -3.78266757949 FDR=0.0 Var=0.07263780555465793 #psms=8]
33 [SMARCAD1 -1.94341647163 FDR=1.08427907229E-6 Var=0.08550152393758327 #psms=4]
34 [ECE2 1.036814925 FDR=0.0197369831178 Var=0.04130271453609074 #psms=3]
35 [IBA57 -0.911784771087 FDR=0.0206146930176 Var=0.042964989805110335 #psms=4]
36 [IGF1R -0.879344881973 FDR=0.0234327104486 Var=0.039576021594976085 #psms=6]
37 [SLC25A16 -0.870042272568 FDR=0.0278610496085 Var=0.04060569687771303 #psms=8]
38 [APP -1.05905086454 FDR=0.029181119344 Var=0.07441823116828218 #psms=4]
Unique to: WT (58)
1 [ADSSL1 0.826000654806 FDR=0.0226347286862 Var=0.01329089975046473 #psms=4]
2 [ITGAV -0.883790655896 FDR=0.0214331639457 Var=0.013036214084941277 #psms=5]
3 [ANKMY2 0.762350816756 FDR=0.0471776204485 Var=0.013237571057764312 #psms=4]
4 [NADSYN1 0.816958717931 FDR=0.0457106468187 Var=0.021391432333857263 #psms=3]
5 [LPCAT1 0.799256632793 FDR=0.0448920288646 Var=0.018187960626560924 #psms=5]
6 [AFG3L2 -0.800170407827 FDR=0.0439306870509 Var=0.009569048828336984 #psms=11]
7 [BCL2L1 1.17632277264 FDR=4.71436313077E-4 Var=0.022999336005284938 #psms=5]
8 [MGEA5 -1.21759143507 FDR=4.06549565402E-4 Var=0.02044596284422332 #psms=5]
9 [ME3 -1.16654082146 FDR=2.95089444336E-4 Var=0.012782344169386698 #psms=21]
10 [RPF2 -1.40638510828 FDR=2.79583078168E-6 Var=0.013132502879267295 #psms=3]
11 [AP2A1,AP2A2,NUDT9 -1.51457317283 FDR=2.2535748919E-6 Var=0.021749691857249544 #psms=9]
12 [WDR4 -1.51457317283 FDR=1.7763874193E-6 Var=0.020462100896901494 #psms=5]
13 [CADM4 -1.51457317283 FDR=1.77320642821E-6 Var=0.020305285730854238 #psms=5]
14 [MAP2K4 -2.12029423372 FDR=1.80712587751E-13 Var=0.019349363811568633 #psms=5]
15 [MESDC2 -2.05889368905 FDR=5.17239584497E-12 Var=0.022576391601792353 #psms=3]
16 [SUSD5 -2.12029423372 FDR=2.04506411805E-11 Var=0.030713298262002326 #psms=4]
17 [EML1 -2.05889368905 FDR=4.65813270309E-11 Var=0.028552778674176292 #psms=6]
18 [TSTA3 -0.90025642728 FDR=0.0269827453504 Var=0.01852573804378209 #psms=4]
19 [APPL2 -0.915935735212 FDR=0.0285072180763 Var=0.021670640450308896 #psms=4]
20 [ACAT1 -0.81964316994 FDR=0.033882589885 Var=0.009281865235680965 #psms=4]
21 [HIST1H1A,HIST1H1C,HIST1H1D,HIST1H1E,HIST1H1T -0.803595269305 FDR=0.0352439626193 Var=0.007443517852178804 #psms=7]
22 [NFX1 -2.39592867633 FDR=0.0 Var=0.02339190321476708 #psms=4]
23 [KIF13B -5.64385618977 FDR=0.0 Var=0.019958078225293618 #psms=3]
24 [NFXL1 -6.64385618977 FDR=0.0 Var=0.020820378487580726 #psms=11]
25 [WDR33 -3.32192809489 FDR=0.0 Var=0.020684095860267624 #psms=7]
26 [WDR77 -2.81514499935 FDR=0.0 Var=0.016561422601349383 #psms=8]
27 [DBN1 -4.64385618977 FDR=0.0 Var=0.01970297263703374 #psms=5]
28 [MYO18A -3.23772920339 FDR=0.0 Var=0.012076962972832474 #psms=5]
29 [FLRT2 2.5993177937 FDR=0.0 Var=0.019164097938888253 #psms=4]
30 [PITHD1 -3.47393118833 FDR=0.0 Var=0.02201530307226255 #psms=6]
31 [OCIAD2 -2.251538767 FDR=6.48711853466E-14 Var=0.02484465740930336 #psms=4]
32 [PARD3B -1.35845397091 FDR=9.81874329042E-5 Var=0.026427483811309983 #psms=6]
33 [NDUFA12 -1.25385174032 FDR=6.63063719208E-5 Var=0.01312347388370167 #psms=3]
34 [PTCD3 -1.35845397091 FDR=3.54221613882E-5 Var=0.020494694356784346 #psms=4]
35 [AHR -1.2960264881 FDR=3.21356407214E-5 Var=0.013651518470162268 #psms=4]
36 [RELA 1.9107326619 FDR=1.39415005856E-10 Var=0.024810339810810763 #psms=4]
37 [ITIH5 1.79486390743 FDR=2.05434069755E-10 Var=0.017452751989503284 #psms=3]
38 [KIAA1429 -1.78587519465 FDR=4.13909610934E-8 Var=0.029281689131265425 #psms=3]
39 [UROD 1.59039935587 FDR=1.01618462978E-7 Var=0.020472771665842993 #psms=5]
40 [TRAPPC1 -1.59946207042 FDR=6.22508645751E-7 Var=0.023500312840072632 #psms=7]
41 [MGST1 -1.18442457114 FDR=7.08821992495E-4 Var=0.020576171425333734 #psms=5]
42 [GORASP2 -1.25059709393 FDR=8.1576702848E-4 Var=0.029858073149872433 #psms=3]
43 [ZFX,ZFY -1.1351492112 FDR=0.00116415677448 Var=0.018314987057104964 #psms=4]
44 [FEN1 1.07724299893 FDR=0.00154096058696 Var=0.020039918523076735 #psms=6]
45 [NUDT5 -1.12029423372 FDR=0.00176049894657 Var=0.01980407710001724 #psms=6]
46 [APTX -1.11528435948 FDR=0.00251179991665 Var=0.02251517157250262 #psms=6]
47 [GPM6A 0.971713600981 FDR=0.00254938181448 Var=0.011564344388696414 #psms=7]
48 [SH3BP4 -1.09150442883 FDR=0.00270804615219 Var=0.020407083135970814 #psms=7]
49 [LRRFIP1 0.569438951328 FDR=0.00312328556834 Var=-0.02616672957897698 #psms=13]
50 [ASF1A 0.917075470227 FDR=0.00352824674342 Var=0.007748705857943978 #psms=3]
51 [SRSF5 -1.05889368905 FDR=0.00469597727262 Var=0.021213023513308896 #psms=4]
52 [SOD2 -0.998931937196 FDR=0.0049956803922 Var=0.014188318976252563 #psms=5]
53 [RALGAPB -1.04791424814 FDR=0.00542438998289 Var=0.02156068770124013 #psms=4]
54 [EEFSEC 1.05658352837 FDR=0.00551060220058 Var=0.030754357376724308 #psms=4]
55 [HSPA1B,HSPA6,HSPA7 0.89089242775 FDR=0.00904135360561 Var=0.012680684762544404 #psms=7]
56 [CRIPT -0.880769646945 FDR=0.0107654431338 Var=0.005877417937576084 #psms=17]
57 [TMLHE -0.885010197436 FDR=0.0118614313666 Var=0.007341741877410148 #psms=11]
58 [SORT1 0.817054063506 FDR=0.017703374771 Var=0.009313537952375847 #psms=5]
Unique to: lacZ (66)
1 [RRAGC,RRAGD -0.75155911826 FDR=0.0143139170616 Var=0.013870808062308553 #psms=7]
2 [SCAF11 -1.02914634566 FDR=0.0136464609437 Var=0.0505331641666931 #psms=6]
3 [GRN -0.735278233864 FDR=0.0135837778971 Var=0.011554300670339848 #psms=8]
4 [CUX1 -0.744840112219 FDR=0.0133658937943 Var=0.01238225820218287 #psms=6]
5 [PDP1 -0.772422634895 FDR=0.0131472972846 Var=0.015309950806677027 #psms=4]
6 [GMPR2 -0.755917038758 FDR=0.0116759411471 Var=0.012561367546993123 #psms=6]
7 [TPD52 0.781177901517 FDR=0.00928606727518 Var=0.014989027947940806 #psms=3]
8 [ACAD9 -0.761410376418 FDR=0.00894193733632 Var=0.011295627404377503 #psms=20]
9 [OCIAD1 0.79831590771 FDR=0.00750435250372 Var=0.015436862143139976 #psms=3]
10 [MATN2 0.742951109966 FDR=0.00739716720197 Var=0.009470073622773746 #psms=6]
11 [VARS -0.785875194647 FDR=0.0465774703642 Var=0.029964785714429884 #psms=4]
12 [HN1 0.649848866677 FDR=0.0371183144687 Var=0.011026271837577085 #psms=6]
13 [UNC13D 0.627911210009 FDR=0.0358640157235 Var=0.008110222852377426 #psms=9]
14 [TIMP3 0.647076544455 FDR=0.0352732515714 Var=0.010188244751021064 #psms=7]
15 [CTNNA1,CTNNA2 -0.81096617561 FDR=0.0345030266688 Var=0.029957286271860957 #psms=4]
16 [EIF2A -0.81096617561 FDR=0.0344593601873 Var=0.030031624820235605 #psms=4]
17 [P4HA1 0.610958007371 FDR=0.0344271636298 Var=0.005742401328415379 #psms=26]
18 [GTF2F1 -1.15200309345 FDR=3.00233945358E-4 Var=0.030811714838504103 #psms=4]
19 [RAB10 -1.15200309345 FDR=2.60613788384E-4 Var=0.0299585766378626 #psms=7]
20 [SLC29A1 1.18269229752 FDR=1.49076537076E-4 Var=0.030886184960375672 #psms=4]
21 [ERBB2IP -1.35845397091 FDR=1.06865216851E-4 Var=0.04604442467194855 #psms=3]
22 [ST13,ST13P4,ST13P5 -1.04386596537 FDR=9.2097223417E-5 Var=0.014823667203193565 #psms=3]
23 [NAT10 -1.05899396632 FDR=7.99898171392E-5 Var=0.015373601774737967 #psms=10]
24 [BCAR1 1.1027128921 FDR=6.225677236E-5 Var=0.019023985747105204 #psms=4]
25 [SORBS3 -0.801929217773 FDR=0.0165976735508 Var=0.021276223824020626 #psms=12]
26 [IPO5 0.706569072947 FDR=0.0166682556061 Var=0.011422823831238656 #psms=3]
27 [SDHB -0.724068533695 FDR=0.0166985680088 Var=0.012254040060412382 #psms=5]
28 [TTC38 -0.819151838995 FDR=0.0167352317621 Var=0.02366910483283462 #psms=5]
29 [TOM1L2 0.723758925654 FDR=0.0185809264294 Var=0.014366209479220243 #psms=5]
30 [SUCLG1 -0.712831932121 FDR=0.0186919598676 Var=0.01177742073521891 #psms=14]
31 [CHPF 0.847996906555 FDR=0.0192643133313 Var=0.03067197556335719 #psms=4]
32 [DAP3 -0.798401311871 FDR=0.0218143348695 Var=0.023598510194547406 #psms=6]
33 [ALDH2 -0.695800138756 FDR=0.0227707704394 Var=0.011328461639395536 #psms=3]
34 [SUPT6H 0.871843648509 FDR=0.0147784461542 Var=0.030461732632053047 #psms=3]
35 [CPSF1 -4.05889368905 FDR=0.0 Var=0.03147672136841231 #psms=8]
36 [PRDX4 -1.86181298864 FDR=0.0 Var=0.015160047381962692 #psms=13]
37 [EPB41,EPB41L1,EPB41L2,EPB41L3 -2.05941464824 FDR=0.0 Var=0.016263339790260546 #psms=4]
38 [GAPDH 2.11511621884 FDR=0.0 Var=0.023782512515894846 #psms=3]
39 [FAF2 -2.55639334852 FDR=0.0 Var=0.02936325923999309 #psms=7]
40 [C1orf52 -2.64385618977 FDR=0.0 Var=0.029892096491323582 #psms=5]
41 [ARMC10 -1.68895387794 FDR=4.00568467285E-13 Var=0.01539409851688918 #psms=4]
42 [COL18A1 0.5807694251 FDR=0.0469464000202 Var=0.004561534931156431 #psms=33]
43 [IDH3B -0.677721092531 FDR=0.0487104561113 Var=0.015377946597920956 #psms=4]
44 [STOML2 -0.704558829567 FDR=0.0488339704372 Var=0.01889670099368666 #psms=3]
45 [REC8 0.672360648434 FDR=0.0320571137596 Var=0.0123373168343153 #psms=3]
46 [PTPRK -0.69235941772 FDR=0.0319465868729 Var=0.013305420132130845 #psms=5]
47 [LSG1 -0.903881828638 FDR=0.00198856556439 Var=0.016600859687012665 #psms=4]
48 [PDHB -0.905986624735 FDR=0.00167566739509 Var=0.015730789608860454 #psms=4]
49 [MME -0.882246732648 FDR=0.00119073125503 Var=0.011832241776933667 #psms=5]
50 [CPOX -0.969535821158 FDR=9.68953069554E-4 Var=0.01906110614259388 #psms=3]
51 [MAT2B -0.95045463796 FDR=7.06036289974E-4 Var=0.0157385035183588 #psms=7]
52 [NSUN2 -1.0892673381 FDR=6.9060394115E-4 Var=0.029417620568434378 #psms=4]
53 [UBA6 -2.32192809489 FDR=0.0 Var=0.02959832292360461 #psms=4]
54 [HBA1 -2.64385618977 FDR=0.0 Var=0.029974361494640184 #psms=3]
55 [FAM102A -5.05889368905 FDR=0.0 Var=0.029295728806327 #psms=4]
56 [UBXN1 -1.43440282415 FDR=1.75206737737E-6 Var=0.031597439503274476 #psms=5]
57 [RAVER1 -1.34876280986 FDR=2.36336443852E-6 Var=0.025759814276783456 #psms=3]
58 [NTPCR 1.37851162325 FDR=2.44531120422E-6 Var=0.029453193995020673 #psms=6]
59 [VTA1 -1.17582225739 FDR=5.43530713462E-6 Var=0.015296249252537029 #psms=3]
60 [CERS2 1.2177622547 FDR=1.26193882132E-5 Var=0.022809875949462193 #psms=5]
61 [MCFD2 -1.02914634566 FDR=0.00216376649941 Var=0.03067935063497734 #psms=3]
62 [SERPINB8,SERPINB9 -0.874451664779 FDR=0.00266852326482 Var=0.015746954042276155 #psms=7]
63 [PPP4R1,PPP4R1L 0.928228490659 FDR=0.00277915829452 Var=0.022933706404734626 #psms=4]
64 [PTMA,SPAG9 -1.0 FDR=0.00289756602505 Var=0.030285766640821186 #psms=3]
65 [RPF1 0.739182360334 FDR=0.00518726903143 Var=0.007219747447187614 #psms=10]
66 [NIFK -0.943416471634 FDR=0.0064878336566 Var=0.029956821928145066 #psms=6]
Contact person:
Salvador Martínez-Bartolomé (salvador at scripps.edu)
Research Associate
The Scripps Research Institute
10550 North Torrey Pines Road
La Jolla, CA 92037
Git-Hub profile