New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Faster covariance and cospectra calculation with einsum #80
Comments
|
thanks, its true that the current implementation is not super optimized. I will look into it, but feel free to open a PR if you feel to. |
|
I agree that the compute cost of the eigenvalue decomposition is currently one of the most important bottlenecks. I have never tried it, but I have been thinking for a while that a However, numpy arrays are not hashable (which is needed for the cache to work), so I can look this up and come back if I have some benchmarks to share. |
|
This speed-up is promissing! Some remarks:
|
|
For |
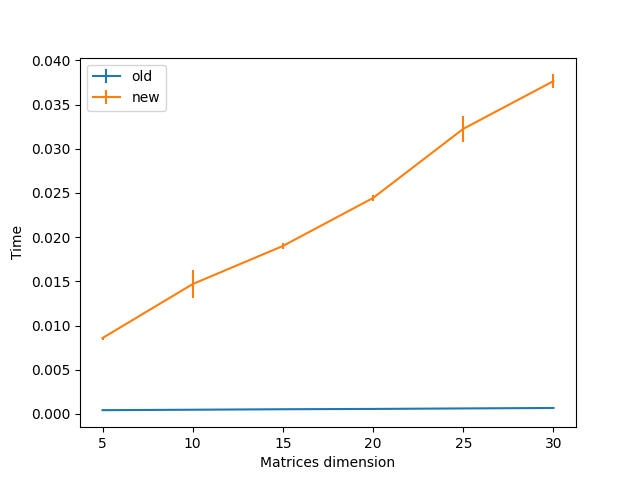
I recently saw that there have been some improvements to the coherence calculation (9a6dbee) and I thought that I would like to propose some further improvements to the covariance and cospectra calculation.
In brief, I have observed that using einsum for these two operations give a speed-up of one order of magnitude. Here is my current code, but let us discuss if and how we could add this to pyriemann:
The text was updated successfully, but these errors were encountered: