New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Numpy-pyccel #942
Comments
I am really surprised that pyccel is so slow that this is counted in seconds! My first instinct is to ask if you are certain that you are running the compiled code and not the python code, or if you are including the translation time in the timeit function? Could I suggest using |
|
The generated code is: The first major difference that I see is that you use are stack arrays in your code while the temporaries are allocatables in the pyccel code. You can ask pyccel to make this optimisation by adding the decorator Beyond this the only differences that I can see aside from the calculation of the norm are algorithmic. In the fortran version the variable In the fortran version the variable In fortran the inner loop has length |
|
The following python file is closer to the original algorithm: It generates the following code: |
|
Hi , Emily , qr_f90 = epyccel(qr_mgs) ERROR at parsing (syntax) stage |
It looks like you need to update your version of pyccel. I think it is complaining about the matmul operator but this now exists in the most up to date version of pyccel (1.3.0). I would suggest updating (unless you are using a mac as there is a compiler bug that will be fixed in version 1.3.1 as soon as #938 is merged) If you can't/don't want to update then the following should also work: |
Feel free to open an issue for this. I think it was left until last as |
|
I update pyccel . |
Haha, that looks much better! I have the following set up: test_qr.py: Where I can therefore run: You can then modify the generated file |
|
In your code you iterate over the last index I have tried making this change as well as manually using I also tried increasing the problem size from 700x700 to 1000x1000. This had the same time ratio so I don't think the wrapper is the bottleneck. Perhaps f2py uses better compiler flags than we do? Or gcc is better at linking than gfortran? (But I think they should both use the same linker in the background) |
|
which optimization flags are you using in f2py? |
My f2py is using the flag Pyccel uses F2PY: Pyccel: (or see I can add the |
According to your document the fastest tool is problem (and compiler) dependant. Assuming you are using a GCC compiler then numba and pythran are both faster in the Laplace case. |
|
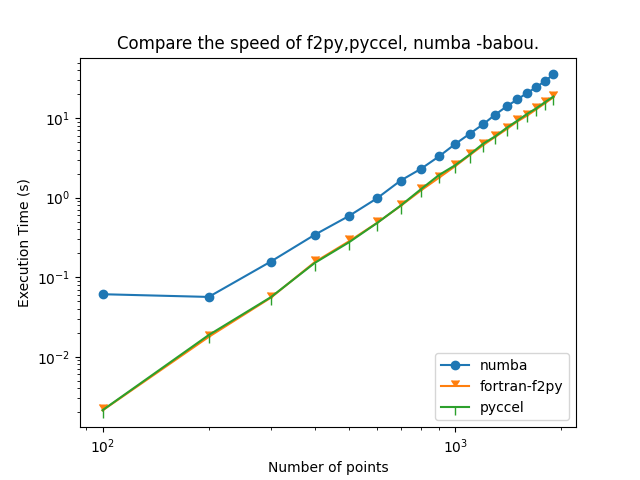
@mohamedlaminebabou Could you redraw your figure, using the branch |
|
@EmilyBourne can you test the pyccel version with the flags |
|
@mohamedlaminebabou you are using the flag |
On my computer I get:
f2py takes approximately 3.2s |
|
@EmilyBourne this is what I have for the flags pyccel version
30.89082655403763
inlined version
9.11238155211322
f2py version
9.146705145947635 |
Is that on the master branch? It's strange that inlining the function would make a difference. The function call itself shouldn't be that costly. The only difference that I can think of is from the declaration of Did you do the inlining manually? Did you copy the function into the bind_c file or did you rewrite the function using What compiler are you using? Mine is |
|
yes I did it manually by replacing the function call with the body of the function |
I can reproduce what you are seeing with this compiler. The behaviour appears to have been improved by gcc between 7.5 and 9.3 although I haven't been able to pinpoint when from the change log |
|
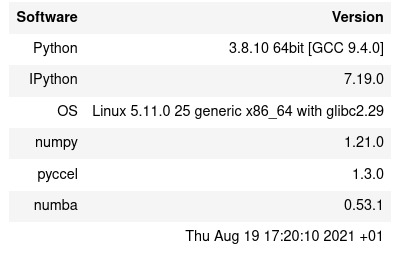
@saidctb @EmilyBourne Thanks for your quick response . Exactly @saidctb when work with the option '-Ofast' and flags "-O3 -funroll-loops -march=native -ffast-math" Indeed, we find that, f2py and pyccel they both have the same speed , but numba fails to progress!. The code used and system info :
|
|
@saidctb When I compile with the previous commands, in jupyter-notebook I have a problem with permision to compile /usr/local/lib/python3.8/dist-packages/pyccel/epyccel.py in /usr/local/lib/python3.8/dist-packages/pyccel/codegen/pipeline.py in /usr/local/lib/python3.8/dist-packages/pyccel/codegen/compiling/compilers.py in /usr/local/lib/python3.8/dist-packages/filelock.py in enter(self) /usr/local/lib/python3.8/dist-packages/filelock.py in acquire(self, timeout, poll_intervall) /usr/local/lib/python3.8/dist-packages/filelock.py in _acquire(self) PermissionError: [Errno 13] Permission denied: '/usr/local/lib/python3.8/dist-packages/pyccel/codegen/compiling/../../compilers.lock' "" |
Please could you create an issue for this. It needs fixing and is unrelated to the current issue.
You can do this using the |
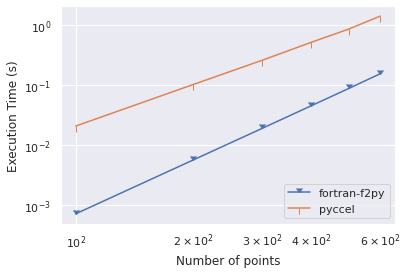

Describe the bug
Hello,
I create a program in fortran and then I transformed it to python and I use Pyccel to accelerate Python functions, but when I use numpy functions my code pyccel is slow than pure fortran , and when I don't use numpy functions pyccel be faster than my fortran code .
So is there a possibility to combine the tow together (fortran code + pyccel ) ?
To Reproduce
Provide code to reproduce the behavior:
Provide the generated code, or the error message:
Expected behavior
A clear and concise description of what you expected to happen.
Language
Please specify which language the python code is translated to (Fortran by default)
Additional context
Add any other context about the problem here.
The text was updated successfully, but these errors were encountered: