PySAL, the Python spatial analysis library, will be changing its package structure.
-
We are changing the module structure to better reflect what you do with the library rather than the academic disciplines the components of the library come from.
-
This also makes the library significantly more maintainable for us, since it reduces the bulk of the library and more evenly distributes the load for maintainers.
-
As an added benefit, we will release these components, called submodules, independently. This lets end users only install components they need, which is helpful for our colleagues in restricted data centers.
-
the main reason we are doing this, though, is so we can implement new features in an easier fashion, maintain existing features more easily, and solicit new contributions and modules with less friction.
Practially speaking, this means that pysal is a single source redistribution of many separately-maintained packages, called submodules. Each of these submodules are available by themselves on PyPI. They are maintained by individuals more closely tied to their code base, and are released on their own schedule. Every six months, the main maintainers of PySAL will collate, test, and re-distribute a stable version of these submodules. The single source release is intended to support our re-packagers, like OSGeoLIVE, Debian, and Conda, as well as most end users. Each subpackage, the individual components available on PyPI, are the locus of development, and are pushed forward (or created anew) by their specific maintainers on their respective repositories. For developers interested in minimizing their dependency requirements, it is possible to depend on each subpackage alone.
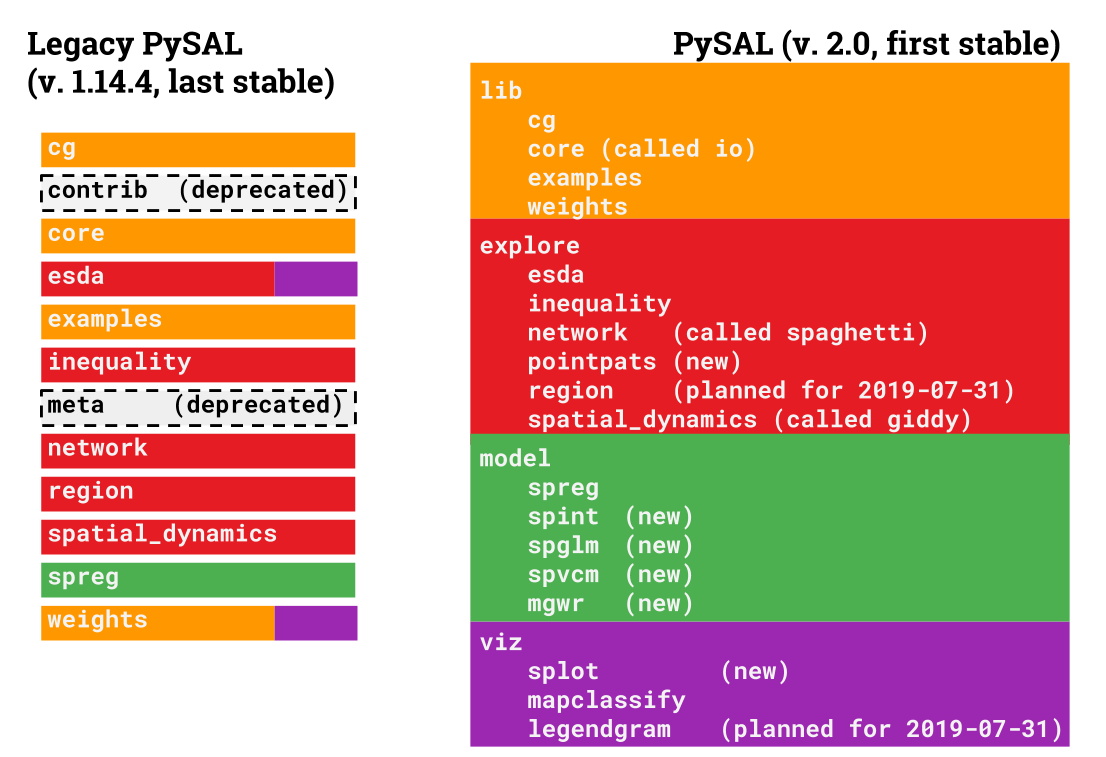
To make this happen, we must re-arrange some existing functionality in the library so that it can be packaged separately. In total, pysal 2.0 will be organized with five main thematic modules with the following functionality:
-
lib: core functionality used by other modules to work with spatial data in Python, including:- the construction of graphs (also known as spatial weights) from spatial data
- computational geometry algorithms:
- alpha shapes
- quadtrees, rTrees, & spherical KDTrees
- methods to construct graphs from
scipyDelaunay triangulation objects - Pure Python data structures to represent planar geometric objects
- pure Python readers/writers for graph/spatial weights files and some spatial data types
-
explore: exploratory spatial data analysis of clusters, hotspots, and spatial outliers, plus spatial statistics on graphs and point patterns. These include- Point pattern analysis of centrography and colocation (
$F$ ,$G$,$J$,$K$ statistics in space) - Pattern analysis of point patterns snapped to a network (
$F$ ,$G$,$K$ statistics on networks) - Analysis of spatial patterning on lattices, including
- univariate Join count statistics
- Moran's
$I$ statistics, including local & bivariate methods - Geary's
$C$ - Getis-Ord
$G$ ,$G^*$ , and local$G$ statistics - General
$\Gamma$ statistics for spatial clustering
- methods to construct & analyze the space-time dynamics and distributions of data, including Markov models and distributional dynamics statistics
- Point pattern analysis of centrography and colocation (
-
model: explicitly spatial modeling tools including:- geographically-weighted regression, a generalized additive spatial model specification for local coefficients
- spatially-correlated variance-components models, a type of Bayesian hierarchical model providing for group-level spatial mixed effects, as well as diagnostics for Bayesian modeling (Geweke, Markov Chain Monte Carlo Standard Error, Potential Scale Reduction Factors)
- Bayesian spatially-varying coefficient process models (e.g. local random effects models)
- Maximum Likelihood spatial econometric models, including:
- mixed regressive-autoregressive (spatial lag) model
- spatially-correlated error models
- spatial regimes/spatial mixture models
- seemingly-unrelated regression models
- combinations of these various effects
- econometric specification testing methods, including spatial Lagrange Multiplier, Anselin-Kelejian, Chow, and Jarque-Bera tests.
viz: methods to visualize and analyze spatial datasets, specifically the output of exploratory spatial statistics.
This document provides a brief overview of what each of the new modules are, how they relate to the last legacy version of pysal, version 1.14.4, and what you need to do in order to keep your code running the way you expect.
The changes in pysal collect together modules that are used for a similar purpose:
Many things are new in pysal 2.0. In order to ensure we can keep making new things easily and maintain what we have, we've released each of the submodules (those underneath model,viz, and explore) as their own python packages on PyPI. We will also release the lib module as its own package, libpysal, on PyPI.
This means that new users can make new submodules by following our submodule contract. Further, contributors should make pull requests to the submodules directly, not to pysal/pysal.
If you want to keep your usual stable pysal dependency, it is sufficient to update your imports according to the mappings we provide in the Module Lookup section. pysal will continue to stick to regular 6-month releases put out by multiple maintainers, with bug-fix releases as needed throughout the year.
This version will still have nightly regression testing run and will be ensured to work with the latest releases of numpy and scipy. If you don't have an urgent need to reduce your dependency size or the availability of PySAL, continuing to depend on pysal directly is the right choice.
If you only use one contained part of pysal, are interested in developing another statistical analysis package that only depends on libpysal, or simply want to keep your build as lean as possible, you can also install only the sub-modules you require, independently of pysal. This is the best option for those of you in restricted analysis environments where every line of code must be vetted by an expert, such as users in restricted data centers conducting academic work.
To preview these changes, install the pysalnext package using pip. If your package works with pysalnext, it should work on pysal 2.0.
All of the sub-packages included in pysalnext contain a significant amount of new functionality, as well as interoperability tools for other packages, such as networkx and geopandas. In addition, most of the old tools from pysal are reorganized. In total, there are 12 distinct packages in pysalnext, with more being added often. These packages are:
libpysal: the core of the library, containing computational geometry, graph construction, and read/write tools.esda: the exploratory spatial data analysis toolkit, containing many statistical functions for characterizing spatial patterns.pointpats: methods and statistical functions to statistically analyze spatial clustering in point patternsspaghetti: methods and statistical functions to analyze geographic networks, including the statistical distribution of points on network topologies.giddy: the geospatial distribution dynamics package, designed to study and characterize how distributions change and relate over space and time.mapclassify: a package with map classification algorithms for cartographysplot: a collection of statistical visualizations for the analysis methods included acrosspysalnextgwr: geographically-weighted regression (both single- and multi-scale)spglm: methods & functions to fit sparse GLMsspint: spatial interaction modelsspreg: spatial econometric regressionspvcm: spatially-correlated variance components models, plus diagnostics and plotting for Bayesian models fit inpysalnext.
There are four main changes that have occurred in pysalnext. From legacy pysal, these are:
-
pysal.contrib: removed due to lack of unittesting and heterogeneous code quality; moved to independent modules where possible. -
pysal.esda.smoothing: removed due to intractable and subtle numerical bugs in the code caused by porting to Python 3. There is an effort to re-implement this in Python, and will be added when/if this effort finishes. -
pysal.region: removed because the new version has significantly more dependencies, includingpulpandscikit-learn. Grab this as a standalone package usingpip install region. -
pysal.meta: removed.
If you'd like to get code from submodules, they usually have a one-to-one replacement. This will be discussed later in the Module Lookup section.
Here is a list of the locations of all of the commonly-used modules in legacy pysal, and where they will move in the next release of pysal.
**To preview these changes, install the pysalnext package using pip. If your package works with pysalnext, it should work on pysal 2.0. **
pysal.cgwill change topysal.lib.cgpysal.weightswill change topysal.lib.weights, and many weights construction classes now havefrom_geodataframemethods that can graphs directly fromgeopandasGeoDataFrames.pysal.openwill change topysal.lib.io.open, and most ofpysal.corewill move topysal.lib.io. Note: usingpysal.lib.io.openfor anything but reading/writing spatial weights matrices/graphs is not advised. Please migrate togeopandas* for all spatial data read/write. Further, note that*WeightsConverterhas also been deprecated; if you need to convert weights, do so manually using sequentialopen* andwritestatements.*pysal.exampleswill change topysal.lib.examples
pysal.spatial_dynamics will change to pysal.explore.giddy
pysal.inequality will change to pysal.explore.inequality
These will mainly move into pysal.explore.esda, except for smoothing and mixture_smoothing (which will be deprecated) and mapclassify, which will move to pysal.viz.mapclassify.
These will move directly into pysal.explore.spaghetti
pysal.inequality has been published as its own package, inequality, and moved to pysal.explore.inequality.
pysal.spreg has been moved wholesale into pysal.model.spreg, which now contains many additional kinds of spatial regression models, including spatial interaction, Bayesian multilevel, and geographically-weighted regression methods.
import pysal
file_handler = pysal.open(pysal.examples.get_path('columbus.dbf'))
data = np.asarray(file_handler.by_col('HOVAL'))becomes:
import geopandas
from pysal.lib import examples
dataframe = geopandas.read_file(examples.get_path('columbus.dbf'))
data = data['HOVAL'].valuesimport pysal
graph = pysal.open(pysal.examples.get_path('columbus.gal')).read()becomes
from pysal.lib import weights, examples
graph = weights.W.from_file(examples.get_path('columbus.gal'))or, building directly on top of the developer-focused libpysal package:
from libpysal import weights, examples
graph = weights.W.from_file(examples.get_path('columbus.gal'))from pysal.esda.mapclassify import Jenks_Caspall
Jenks_Caspall(your_data).ybbecomes
from pysal.viz import mapclassify
labels = mapclassify.Jenks_Caspall(features).ybOr, built directly on top of the developer-focused package mapclassify, which may have newer features in the future:
import mapclassify
labels = mapclassify.Jenks_Caspall(features).ybimport pysal
file_handler = pysal.open(pysal.examples.get_path('columbus.dbf'))
y = np.asarray(file_handler.by_col('HOVAL'))
X = file_handler.by_col_array(['CRIME', 'INC'])
graph = pysal.queen_from_shapefile(pysal.examples.get_path('columbus.shp'))
model = pysal.spreg.ML_Lag(y,X, w=graph,
name_x = ['CRIME', 'INC'],
name_y = 'HOVAL')becomes
from pysal.model import spreg
from pysal.lib import weights, examples
import geopandas
dataframe = geopandas.read_file(examples.get_path("columbus.dbf"))
graph = weights.Queen.from_dataframe(dataframe) # Queen.from_shapefile also supported
model = spreg.ML_Lag(dataframe[['HOVAL']].values,
dataframe[['CRIME', 'INC']].values,
name_x = ['CRIME', 'INC'], name_y = 'HOVAL')Or, building on top of the standalone spreg package, which may have new bugfixes or compatibility options in the future:
from pysal.model import spreg
from pysal.lib import weights, examples
import geopandas
dataframe = geopandas.read_file(examples.get_path("columbus.dbf"))
graph = weights.Queen.from_dataframe(dataframe)
model = spreg.ML_Lag(dataframe[['HOVAL']].values,
dataframe[['CRIME', 'INC']].values,
name_x = ['CRIME', 'INC'], name_y = 'HOVAL')
model2 = spreg.ML_Lag.from_formula('HOVAL ~ CRIME + INC',
data=dataframe)import pysal
file_handler = pysal.open(pysal.examples.get_path('columbus.dbf'))
y = np.asarray(file_handler.by_col('HOVAL'))
graph = pysal.open(pysal.examples.get_path('columbus.gal')).read()
moran_stat = pysal.Moran(y,graph)
print(moran_stat.I, moran_stat.p_z_sim)becomes
from pysal.explore import esda
from pysal.lib import weights, examples
import geopandas
dataframe = geopandas.read_file(examples.get_path("columbus.dbf"))
graph = weights.Queen.from_dataframe(dataframe)
moran_stat = esda.Moran(dataframe['HOVAL'], graph)
print(moran_stat.I, moran_stat.p_z_sim)or, building directly off of the developer-focused package esda , which may have features not yet available in pysal itself:
import esda
from pysal.lib import weights, examples
import geopandas
dataframe = geopandas.read_file(examples.get_path("columbus.dbf"))
graph = weights.Queen.from_dataframe(dataframe)
moran_stat = esda.Moran(dataframe['HOVAL'], graph, fancy_new_option=True)
print(moran_stat.I, moran_stat.p_z_sim)This is not recommended.
For a longer change window, feel free to import pysal._legacy as pysal. We urge you to not do this, since we plan on deprecating this as well. If you can make the changes described above, you will have a much more stable and future-proof API. We feel these changes are reasonable and will greatly enhance how easy it is for us to maintain pysal and move new functionality forward.