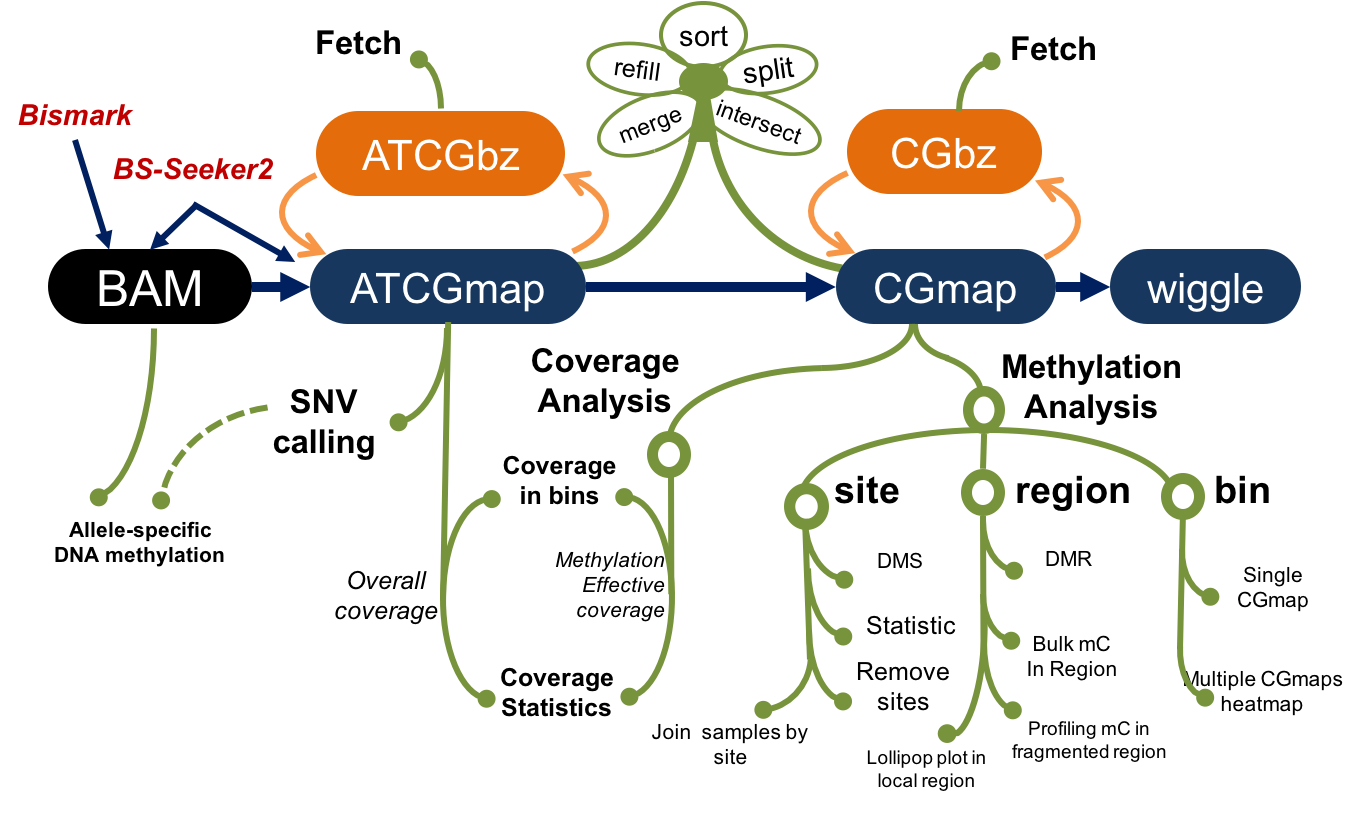
Command-line toolset for advanced analysis of bisulfite sequencing data.
Citation: Guo, W. et al. CGmapTools improves the precision of heterozygous SNV calls and supports allele-specific methylation detection and visualization in bisulfite-sequencing data. Bioinformatics 34, 381–387 (2018).
- Online tutorial: https://cgmaptools.github.io
- For primary data processing and mapping of bisulfite sequencing data, particularly at single-cell resolution, we highly recommend our recently developed tool scBS-map (click to jump to scBS-map github page).