-
Notifications
You must be signed in to change notification settings - Fork 514
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Fix RF regression performance #3845
Conversation
|
Thank you for the optimizations, Rory! Yes, the usage of mean for MAE is one of the approximations in this code! However, I think we should still keep the MAE code because we could potentially use Jensen's inequality to help approximate the computation of median. |
|
The iterative median approach seems interesting, but all of the MAE approaches seem to be fairly challenging I think? How important is MAE for RF now? I have not heard of folks using it actively, perhaps because the existing sklearn implementation takes far longer than MSE. (Basically if you search for RandomForestRegressor and MAE almost all of the hits are complainta about perf.) A fast implementation could be interesting, but I would like to consider how widely used it is before we spend too much time optimizing. |
|
Benchmarks comparing the PR with branch-0.20 and sklearn
|
Co-authored-by: Philip Hyunsu Cho <chohyu01@cs.washington.edu>
|
Dropping MAE support looks very dangerous to me as it give one reason to not use cuml. People won't mix cuml and non cuml models easily, and MAE is used in the industry. I have used it. I google parallel median computation and found a number of papers. This one in particular discusses median computation on GPU https://arxiv.org/pdf/1104.2732.pdf And if implementing one of these is too costly, then keeping the existing approximation would be better than removing it altogether IMHO. |
|
@jfpuget It's not just median calculation, it's streaming median calculation. Full implementation is off the table right now. I believe current MAE is incorrect from a user perspective, so we would just be giving the appearance of supporting it. That seems misleading to me. Here is some evidence: Let the labels be a beta distribution with a clear separation between the mean (red) and median (green): Build a depth 6 decision tree on a random X matrix, the MAE is as follows: cuml's MAE completely tracks MSE. Put another way, cuml never actually had the MAE feature to begin with. Here is my script: from cuml import RandomForestRegressor as cuRF
from sklearn.ensemble import RandomForestRegressor as sklRF
from sklearn.datasets import make_regression
from sklearn.metrics import mean_absolute_error
import numpy as np
import pandas as pd
from scipy.stats import beta
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
matplotlib.use("Agg")
sns.set()
n = 1000
X = np.full((1000, 1), 1.0, dtype=np.float32)
X = np.random.random((1000, 1))
a, b = 2.31, 0.627
y = beta.rvs(a, b, size=n)
print(y.mean())
print(np.median(y))
sns.displot(y, kind="kde")
plt.axvline(y.mean(),color='r')
plt.axvline(np.median(y),color='g')
plt.savefig("beta.png")
plt.clf()
rs = np.random.RandomState(92)
df = pd.DataFrame(columns=["algorithm", "accuracy", "depth"])
d=6
n_repeats = 1
bootstrap = False
max_samples = 1.0
max_features = 1.0
n_estimators = 1
n_bins = min(X.shape[0], 128)
clf = sklRF(
n_estimators=n_estimators,
max_depth=d,
random_state=rs,
max_features=max_features,
bootstrap=bootstrap,
max_samples=max_samples if max_samples < 1.0 else None,
criterion="mae",
)
clf.fit(X, y)
pred = clf.predict(X)
cu_clf = cuRF(
n_estimators=n_estimators,
max_depth=d,
random_state=rs.randint(0, 1 << 32),
n_bins=n_bins,
max_features=max_features,
bootstrap=bootstrap,
max_samples=max_samples,
split_criterion=3,
use_experimental_backend=True,
)
cu_clf.fit(X, y)
cu_pred = cu_clf.predict(X, predict_model="CPU")
cu_clf_mse = cuRF(
n_estimators=n_estimators,
max_depth=d,
random_state=rs.randint(0, 1 << 32),
n_bins=n_bins,
max_features=max_features,
bootstrap=bootstrap,
max_samples=max_samples,
split_criterion=2,
use_experimental_backend=True,
)
cu_clf_mse.fit(X, y)
cu_mse_pred = cu_clf_mse.predict(X, predict_model="CPU")
skl_accuracy = mean_absolute_error(y, pred)
cu_accuracy = mean_absolute_error(y, cu_pred)
cu_mse_accuracy = mean_absolute_error(y, cu_mse_pred)
df = df.append(
{"algorithm": "cuml_mae", "accuracy": cu_accuracy, "depth": d},
ignore_index=True,
)
df = df.append(
{"algorithm": "cuml_mse", "accuracy": cu_accuracy, "depth": d},
ignore_index=True,
)
df = df.append(
{"algorithm": "sklearn", "accuracy": skl_accuracy, "depth": d},
ignore_index=True,
)
print(df)
sns.barplot(data=df, x="algorithm", y="accuracy")
plt.ylabel("MAE")
plt.savefig("beta_rf.png") |
|
I think the framing of this question has been incorrect. We don't have MAE today, so we can't really ask "should we keep MAE?" We have an incorrect metric (essentially MSE-but-slower) masquerading as MAE today. Even without the 5x+ speedup in regression from this PR, I think it would be wise to remove this incorrectly-labeled metric to avoid user confusion, as the current situation (silently using the completely wrong metric) is very user hostile. The question of - "should we work on a proper MAE in the future?" is separate but still interesting. This is absolutely something we can consider. It will be costly in terms of engineering work and will still be much slower than MSE, so we'd look for detailed user use cases to justify it and roadmap it, as with all features. |
|
I'd like to provide a slightly different perspective... I'm perfectly fine removing MAE for now, as long as we can promise to spend time in our next releases to add it back without asking for justification. From end users' perspective, MAE (for better or worse, whether it was completely incorrect or useless or not) was already there. Hence, IMO, removing it and then asking for justification from the users to put it back, is not a good idea. |
|
Fair enough, I agree MAE isn't there today, hence this would not remove MAE. But we need to support MAE, hence planning for tis support looks right to me. |
|
@RAMitchell there seems to be a couple googletests that need to be updated to "drop" MAE: [ RUN ] RfRegressorTests/RfRegressorTestF.Fit/3
unknown file: Failure
C++ exception with description "exception occured! file=../src/decisiontree/decisiontree.cu line=98: MAE not supported.
Obtained 16 stack frames
#0 in ./test/ml(_ZN4raft9exception18collect_call_stackEv+0x3b) [0x4c56bb]
#1 in ./test/ml(_ZN4raft9exceptionC2ENSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEEE+0x6d) [0x4c5d4d]
#2 in /workspace/ci/artifacts/cuml/cpu/conda_work/cpp/build/libcuml++.so(+0x1a26be) [0x7ff32e0496be]
#3 in /workspace/ci/artifacts/cuml/cpu/conda_work/cpp/build/libcuml++.so(_ZN2ML14validity_checkENS_9RF_paramsE+0x326) [0x7ff32e4c6d16]
#4 in /workspace/ci/artifacts/cuml/cpu/conda_work/cpp/build/libcuml++.so(_ZN2ML11rfRegressorIfEC2ENS_9RF_paramsE+0x61) [0x7ff32e4d5f81]
#5 in /workspace/ci/artifacts/cuml/cpu/conda_work/cpp/build/libcuml++.so(_ZN2ML3fitERKN4raft8handle_tERPNS_20RandomForestMetaDataIffEEPfiiS8_NS_9RF_paramsEi+0x288) [0x7ff32e4cf948]
#6 in ./test/ml() [0x6e4c71]
#7 in /opt/conda/envs/rapids/lib/libgtest.so(_ZN7testing8internal35HandleExceptionsInMethodIfSupportedINS_4TestEvEET0_PT_MS4_FS3_vEPKc+0x4e) [0x7ff37c17398e]
#8 in /opt/conda/envs/rapids/lib/libgtest.so(_ZN7testing4Test3RunEv+0x64) [0x7ff37c173b64]
#9 in /opt/conda/envs/rapids/lib/libgtest.so(_ZN7testing8TestInfo3RunEv+0x13f) [0x7ff37c173f0f]
#10 in /opt/conda/envs/rapids/lib/libgtest.so(_ZN7testing9TestSuite3RunEv+0x106) [0x7ff37c174036]
#11 in /opt/conda/envs/rapids/lib/libgtest.so(_ZN7testing8internal12UnitTestImpl11RunAllTestsEv+0x4dc) [0x7ff37c1745ec]
#12 in /opt/conda/envs/rapids/lib/libgtest.so(_ZN7testing8UnitTest3RunEv+0xd9) [0x7ff37c174859]
#13 in /opt/conda/envs/rapids/lib/libgtest_main.so(main+0x3f) [0x7ff37c12307f]
#14 in /lib/x86_64-linux-gnu/libc.so.6(__libc_start_main+0xf0) [0x7ff32d245840]
#15 in ./test/ml() [0x4b6029]
" thrown in SetUp().
[ FAILED ] RfRegressorTests/RfRegressorTestF.Fit/3, where GetParam() = (1 ms)(posting from the logs to help with the parsing of why CI is failing 🙂 ) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
LGTM. I especially took careful attention to the MSE gain calculation, and everything works out.
Codecov Report
@@ Coverage Diff @@
## branch-21.06 #3845 +/- ##
===============================================
Coverage ? 85.41%
===============================================
Files ? 227
Lines ? 17317
Branches ? 0
===============================================
Hits ? 14791
Misses ? 2526
Partials ? 0
Flags with carried forward coverage won't be shown. Click here to find out more. Continue to review full report at Codecov.
|
| @@ -610,15 +605,10 @@ __global__ void computeSplitRegressionKernel( | |||
| // transfer from global to smem | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
gs.sync() on line 603 is not required anymore as there is no second pass. This change plus the single block bypass trick I implemented as part of #3818 performed 10-12% better in my testing.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Approving on behalf of cpp codeowners based on reviews by Philip and Vinay (thank you guys!)
|
@JohnZed the only thing remaining is the approval from you (or any other python codeowners) and we are good to merge! |
|
@gpucibot merge |
…ate old-backend. (#3872) * This PR follows #3845 and resolves #3520 * Makes new-backend default for regression tasks. Now, for both classification and regression tasks, experimental-backend (new-backend) is better than old 😀 * Adds deprecation warning when using old-backend in the C++ DecisionTree layer so that the warning reflects for the decision trees C++ API too. * Sets default `n_bins` to 128 * Some docs update * Some python tests update Authors: - Venkat (https://github.com/venkywonka) - Rory Mitchell (https://github.com/RAMitchell) Approvers: - Rory Mitchell (https://github.com/RAMitchell) - Thejaswi. N. S (https://github.com/teju85) - Philip Hyunsu Cho (https://github.com/hcho3) - Dante Gama Dessavre (https://github.com/dantegd) URL: #3872
This PR rewrites the mean squared error objective. Mean squared error is much easier when factored mathematically into a slightly different form. This should bring regression performance in line with classification. I've also removed the MAE objective as its not correct. This can be seen from the fact that leaf predictions with MAE use the mean, where the correct minimiser is the median. Also see sklearns implementation, where streaming median calculations are required: https://github.com/scikit-learn/scikit-learn/blob/de1262c35e2aa4ee062d050281ee576ce9e35c94/sklearn/tree/_criterion.pyx#L976. Implementing this correctly for GPU would be very challenging. Performance before: 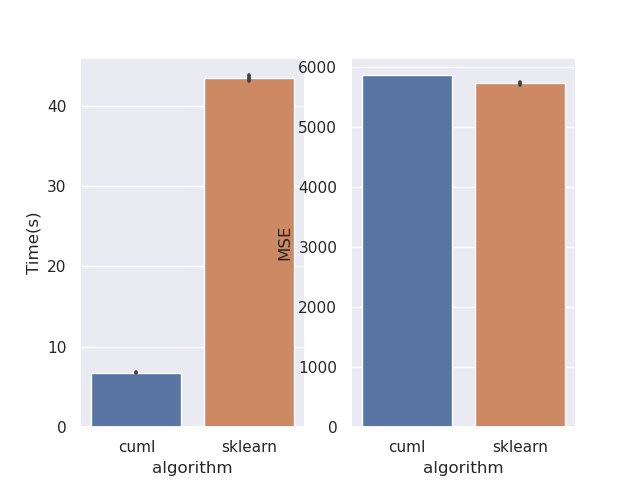 After: 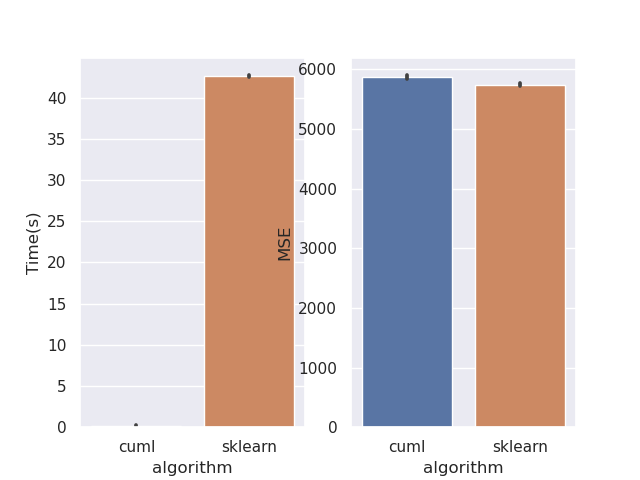 Script: ```python from cuml import RandomForestRegressor as cuRF from sklearn.ensemble import RandomForestRegressor as sklRF from sklearn.datasets import make_regression from sklearn.metrics import mean_squared_error import numpy as np import pandas as pd import matplotlib import matplotlib.pyplot as plt import seaborn as sns import time matplotlib.use("Agg") sns.set() X, y = make_regression(n_samples=100000, random_state=0) X = X.astype(np.float32) y = y.astype(np.float32) rs = np.random.RandomState(92) df = pd.DataFrame(columns=["algorithm", "Time(s)", "MSE"]) d = 10 n_repeats = 5 bootstrap = False max_samples = 1.0 max_features = 0.5 n_estimators = 10 n_bins = min(X.shape[0], 128) for _ in range(n_repeats): clf = sklRF( n_estimators=n_estimators, max_depth=d, random_state=rs, max_features=max_features, bootstrap=bootstrap, max_samples=max_samples if max_samples < 1.0 else None, ) start = time.perf_counter() clf.fit(X, y) skl_time = time.perf_counter() - start pred = clf.predict(X) cu_clf = cuRF( n_estimators=n_estimators, max_depth=d, random_state=rs.randint(0, 1 << 32), n_bins=n_bins, max_features=max_features, bootstrap=bootstrap, max_samples=max_samples, use_experimental_backend=True, ) start = time.perf_counter() cu_clf.fit(X, y) cu_time = time.perf_counter() - start cu_pred = cu_clf.predict(X, predict_model="CPU") df = df.append( { "algorithm": "cuml", "Time(s)": cu_time, "MSE": mean_squared_error(y, cu_pred), }, ignore_index=True, ) df = df.append( { "algorithm": "sklearn", "Time(s)": skl_time, "MSE": mean_squared_error(y, pred), }, ignore_index=True, ) print(df) fig, ax = plt.subplots(1, 2) sns.barplot(data=df, x="algorithm", y="Time(s)", ax=ax[0]) sns.barplot(data=df, x="algorithm", y="MSE", ax=ax[1]) plt.savefig("rf_regression_perf_fix.png") ``` Authors: - Rory Mitchell (https://github.com/RAMitchell) Approvers: - Philip Hyunsu Cho (https://github.com/hcho3) - Thejaswi. N. S (https://github.com/teju85) - John Zedlewski (https://github.com/JohnZed) URL: rapidsai#3845
…ate old-backend. (rapidsai#3872) * This PR follows rapidsai#3845 and resolves rapidsai#3520 * Makes new-backend default for regression tasks. Now, for both classification and regression tasks, experimental-backend (new-backend) is better than old 😀 * Adds deprecation warning when using old-backend in the C++ DecisionTree layer so that the warning reflects for the decision trees C++ API too. * Sets default `n_bins` to 128 * Some docs update * Some python tests update Authors: - Venkat (https://github.com/venkywonka) - Rory Mitchell (https://github.com/RAMitchell) Approvers: - Rory Mitchell (https://github.com/RAMitchell) - Thejaswi. N. S (https://github.com/teju85) - Philip Hyunsu Cho (https://github.com/hcho3) - Dante Gama Dessavre (https://github.com/dantegd) URL: rapidsai#3872





This PR rewrites the mean squared error objective. Mean squared error is much easier when factored mathematically into a slightly different form. This should bring regression performance in line with classification.
I've also removed the MAE objective as its not correct. This can be seen from the fact that leaf predictions with MAE use the mean, where the correct minimiser is the median. Also see sklearns implementation, where streaming median calculations are required: https://github.com/scikit-learn/scikit-learn/blob/de1262c35e2aa4ee062d050281ee576ce9e35c94/sklearn/tree/_criterion.pyx#L976.
Implementing this correctly for GPU would be very challenging.
Performance before:


After:
Script: