You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
If you are using conda, which channel did you install the rdkit from? rdkit
Description:
Ran into a few strange behaviors (at least relative to what I would expect) when trying to code up a substitution reaction. Minimal examples reproduced below:
For all examples the reactant of interest is Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
We begin by testing the simple bimolecular substitution reaction, with inversion of stereochemistry at the reaction center:
The result is 'CCS[C@@H](C)c1ccccc1' (shown below)
However, this is actually not the expected inverted product. The reason for this I believe is due to the change of the order of the SMILES. The carbon indeed changes from [C@] to [C@@]; however, since the canonicalization changes the bond we look down to define clockwise/counter-clockwise, no true inversion occurs. Ultimately, I did not see an easy way to fix this problem (though I welcome suggestions!), so started testing a unimolecular transformation, as shown below.
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:2]([#6:1])[Br:3]>>[C@@:2]([#6:1])[S:3]CC')
prod = rxn.RunReactants(
(
reactant,
)
)
print(len(prod))
print([Chem.MolToSmiles(x[0]) for x in prod])
This example, produces both stereoisomers:
and
I understand that my RXN SMARTS is likely overly general, since '[C@:2]([#6:1])[Br:3]' may be matched in two different ways to the reactant. However, either way it matches, shouldn't the product simply invert the stereochemistry of the original match? It is a bit unclear to me why the configuration of the reactant is retained.
Investigating this behavior, I tried the less general rxn below
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:2]([C:1])[Br:3]>>[C@@:2]([C:1])[S:3]CC')
prod = rxn.RunReactants(
(
reactant,
)
)
print(len(prod))
print([Chem.MolToSmiles(x[0]) for x in prod])
This actually only produces the expected product with inversion of stereochemistry:
Lastly, I tried to isolate the behavior of the aromatic carbon causing troubles:
reactant = Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')
rxn = AllChem.ReactionFromSmarts('[C@:2]([c:1])[Br:3]>>[C@@:2]([c:1])[S:3]CC')
prod = rxn.RunReactants(
(
reactant,
)
)
print(len(prod))
print([Chem.MolToSmiles(x[0]) for x in prod])
This reaction only produces the unexpected product with retention.
I expect that an issue may arise because the original SMILES of the reactant, ''C[C@H](Br)c1ccccc1' is defining counter-clockwise with respect to looking down the non-aromatic CC bond, and not the cC bond.
However, I am pretty confused in general, so would really appreciate any clarification. I apologize for the lengthy issue, but hopefully the reproducible examples are of help.
The text was updated successfully, but these errors were encountered:
Thanks for that detailed and well described bug report!
There's definitely something odd/unexpected going on here that I'm going to have to dig into (it's not a trivial topic) in order to understand/explain.
* add tests for the problem
* more testing (still no fix)
* Fixes#2891
Try to be more robust w.r.t. atom reordering in input SMILES
* better handling of differing numbers of bonds between reactants and products
all tests now passing
* update the rdkit book with more details about chirality in reactions
* changes in response to review
@mc-robinson : I believe we have this fixed on master now. It'll be in the 2020.09.1 release (coming in mid-October).
Here are the new results for a couple of your examples:
Description:
Ran into a few strange behaviors (at least relative to what I would expect) when trying to code up a substitution reaction. Minimal examples reproduced below:
For all examples the reactant of interest is
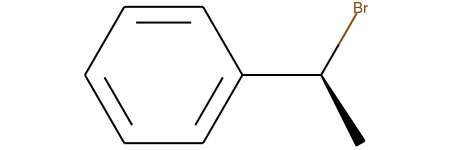
Chem.MolFromSmiles('C[C@H](Br)c1ccccc1')We begin by testing the simple bimolecular substitution reaction, with inversion of stereochemistry at the reaction center:
The result is
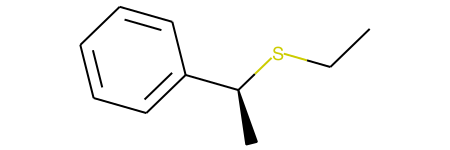
'CCS[C@@H](C)c1ccccc1'(shown below)However, this is actually not the expected inverted product. The reason for this I believe is due to the change of the order of the SMILES. The carbon indeed changes from [C@] to [C@@]; however, since the canonicalization changes the bond we look down to define clockwise/counter-clockwise, no true inversion occurs. Ultimately, I did not see an easy way to fix this problem (though I welcome suggestions!), so started testing a unimolecular transformation, as shown below.
This example, produces both stereoisomers:
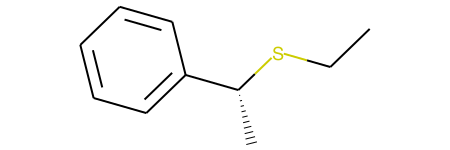
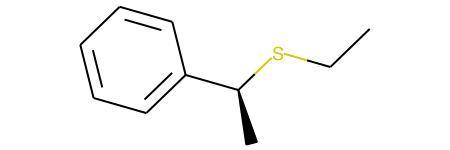
and
I understand that my RXN SMARTS is likely overly general, since
'[C@:2]([#6:1])[Br:3]'may be matched in two different ways to the reactant. However, either way it matches, shouldn't the product simply invert the stereochemistry of the original match? It is a bit unclear to me why the configuration of the reactant is retained.Investigating this behavior, I tried the less general rxn below
This actually only produces the expected product with inversion of stereochemistry:
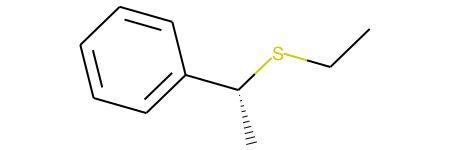
Lastly, I tried to isolate the behavior of the aromatic carbon causing troubles:
This reaction only produces the unexpected product with retention.
I expect that an issue may arise because the original SMILES of the reactant,
''C[C@H](Br)c1ccccc1'is defining counter-clockwise with respect to looking down the non-aromatic CC bond, and not the cC bond.However, I am pretty confused in general, so would really appreciate any clarification. I apologize for the lengthy issue, but hopefully the reproducible examples are of help.
The text was updated successfully, but these errors were encountered: