We provide the official PyTorch implementation of the paper titled Semi-supervised Brain Tumor Segmentation Using Diffusion Models by Ahmed Alshenoudy, Bertram Sabrowsky-Hirsch, Stefan Thumfart, Michael Giretzlehner and Erich Kobler.
Our implementation is based on Label-Efficient Semantic Segmentation with Diffusion Models, where we also employ Improved Denoising Diffusion Probabilistic Models. Various core functions were heavily influenced from nnUNet V1 as well.
In this paper, we leverage learned visual representations from diffusion models for the challenging task of brain tumor segmentation. We compare the segmentation performance against a supervised baseline over a varying degree of training samples. For the downstream segmentation task, we used pixel-level classifiers and additionally proposed the fine-tuning of the noise predictor network of the diffusion model. Our results show that, with less than 20 training samples, all methods outperform the supervised baseline across all tumor regions. We also provide a practical use-case where we automatically annotate tumor regions across different axial slices within the same patient, with very limited supervision.
We evaluate the presented approach on the Brain Tumor Segmentation (BraTS) 2021 data. Axial slices were extracted from the original 3D MR sequences, while stratifying longitudinal slicing locations to increase the proportion of slices containing a segmented tumor. All slices were normalized and down-sampled to (128, 128). This resulted in a dataset consisting of 8,757 slices, of which 8,000 were used for testing and the 757 remaining slices were used as a training pool to sample various training datasets from for our experiments.
- Our trained diffusion model and a small batch of generated samples can be found here. We visualize a batch of generated mpMRI samples from the fully trained model in the figure below. This is to visually assess how well the model is able to approximate the dataset distribution.
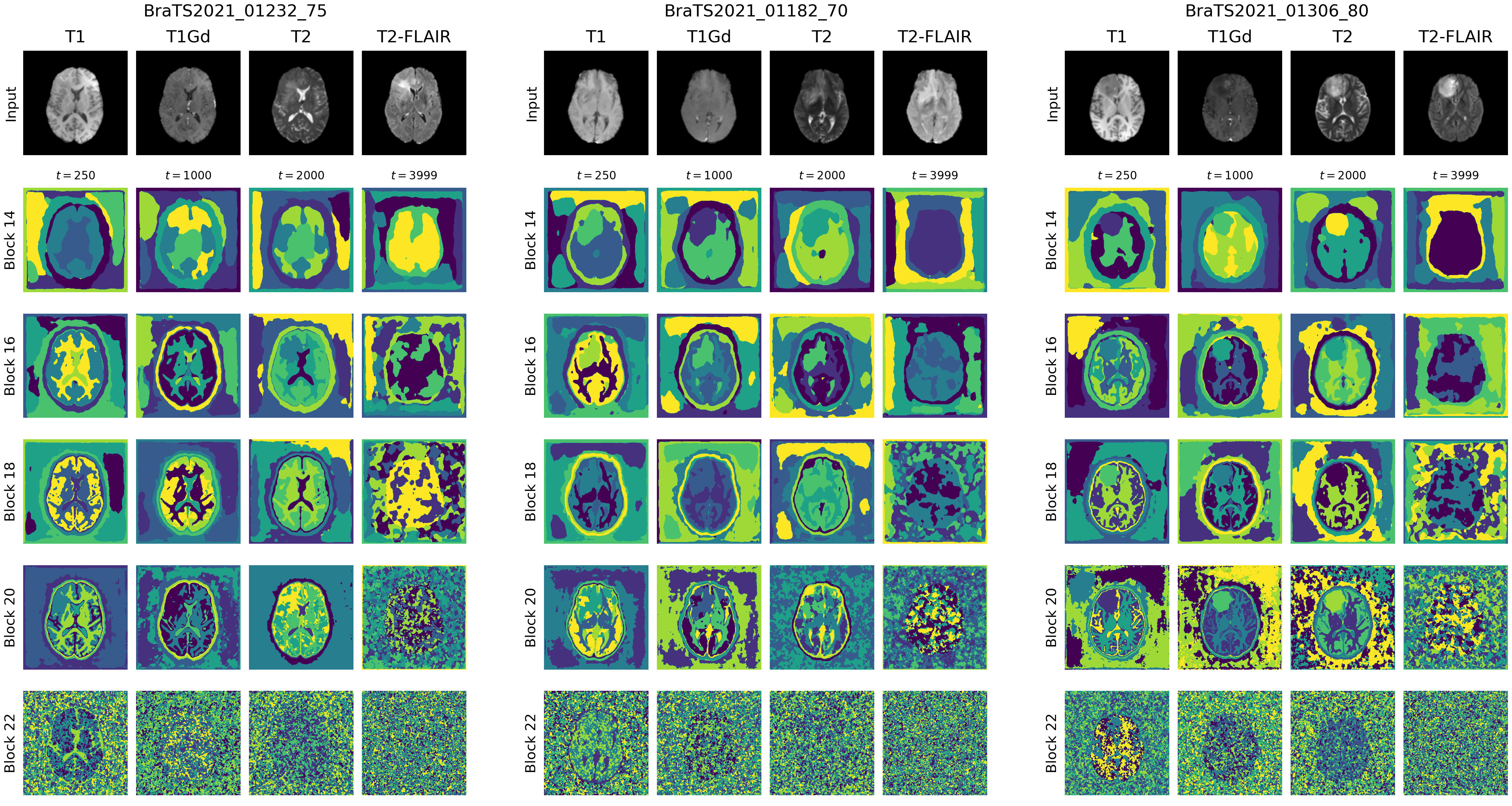
- Extracted visual representations for sample inputs are visualized in Fig. 2, our results align with the original paper, where earlier layer capture high-level features and later layers capture more detailed features while also becoming noisier.
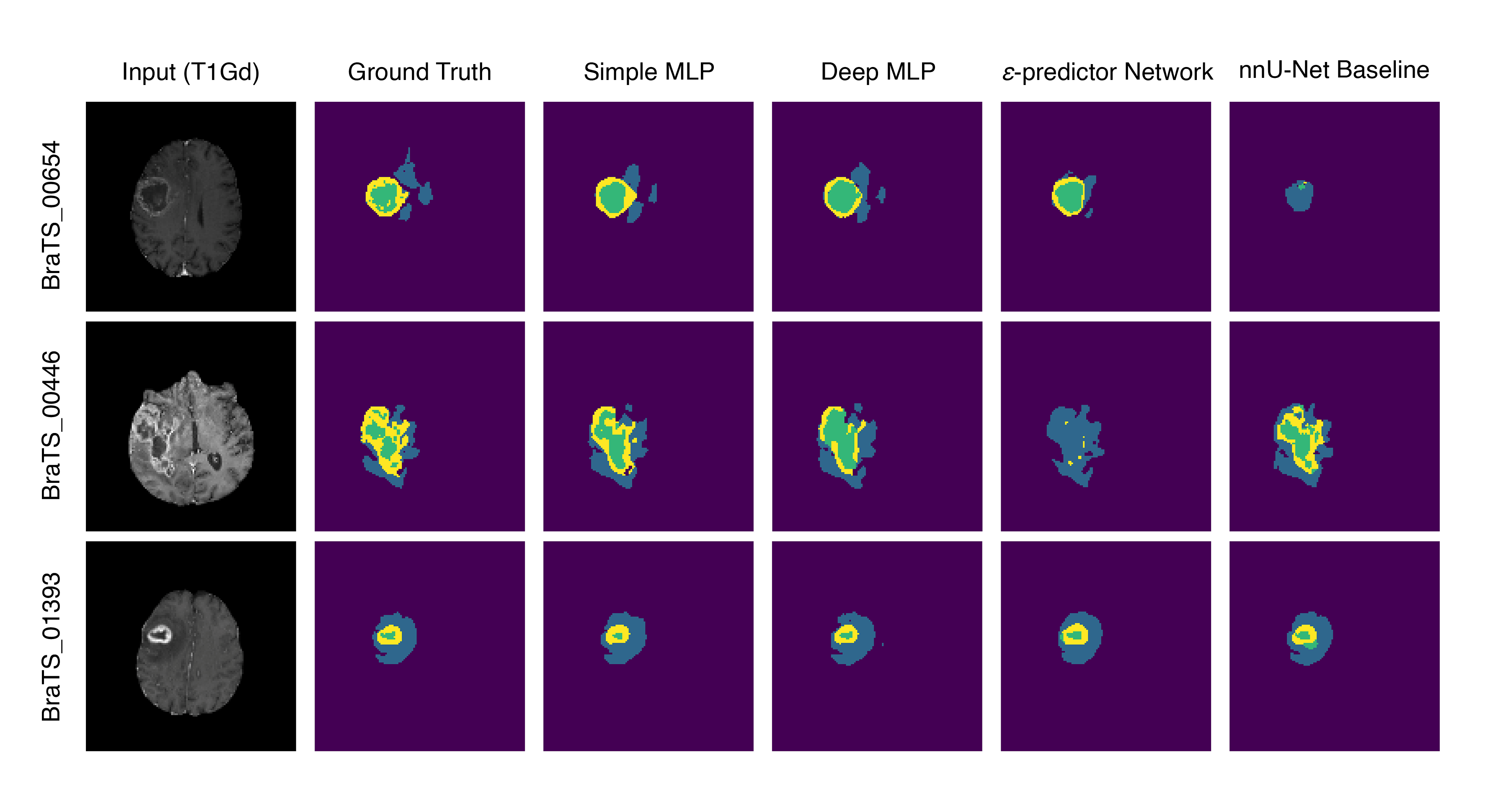
- Sample predictions of different tumor regions are visualized in Fig. 3, a comparison between two pixel-level classifier architectures, the fine-tuned noise predictor network and the nnU-Net supervised baseline.
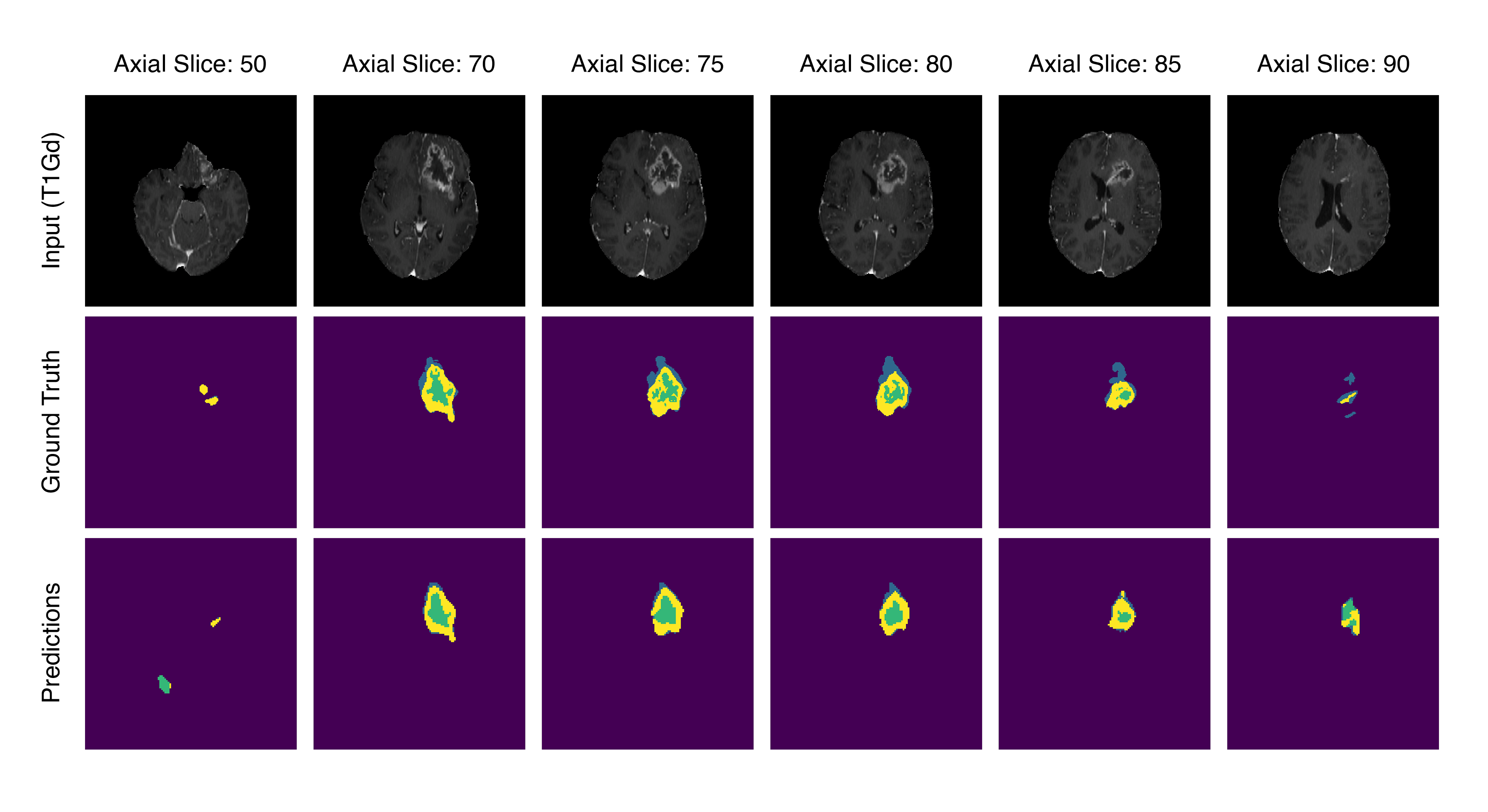
- A sample practical use-case, where a few tumor containing axial slices from the same patient are used to train the downstream pixel-level classifier to produce segmentation maps for the remaining slices within the same volume. Performance is very good for slices that contain larger sections of the tumor, while a significant drop in performance is observed for out of distribution slices that were not used for training the downstream model or the diffusion model itself.
If you find this codebase useful for your research, we would appreciate citing the following conference paper:
@InProceedings{braintumor_ddpm2023,
author={Alshenoudy, Ahmed and Sabrowsky-Hirsch, Bertram and Thumfart, Stefan and Giretzlehner, Michael and Kobler, Erich},
editor={Maglogiannis, Ilias and Iliadis, Lazaros and MacIntyre, John and Dominguez, Manuel},
title={Semi-supervised Brain Tumor Segmentation Using Diffusion Models},
booktitle={Artificial Intelligence Applications and Innovations},
year={2023},
publisher={Springer Nature Switzerland},
address={Cham},
pages={314--325},
isbn={978-3-031-34111-3}.
doi={10.1007/978-3-031-34111-3_27}
}
This project is financed by research subsidies granted by the government of Upper Austria. RISC Software GmbH is Member of UAR (Upper Austrian Research) Innovation Network.
- Tutorial notebook on how to use the pipeline for segmentation
- Addition of Deep/Wide MLP architecture and normal initialization
- Requirements file notes:
- Numpy (< 1.24.0) due to conflict with MedPy (used for HD95 metric)
- Introduced anti-alias parameter in torchvision
- MPI, used for
dist_utilsin improved diffusion