https://pymonnto.readthedocs.io/
The "Python modular neural network toolbox" allows you to create different Neuron-Groups, define their Behavior and connect them with Synapse-Groups.
With this Simulator you can create all kinds of biological plausible networks, which, for example, mimic the learning mechanisms of cortical structures.
PymoNNto requires Python3 and can be installen via pip with the following command:
pip install PymoNNto
If you want to extend the PymoNNto code you can also clone the git repository and manually install the packages defined in requirements.txt
The following code creates a network of 100 neurons with recurrent connections and simulates them for 1000 iterations. What is still missing are some behavior modules. This modules have to be passed to the NeuronGrop to definde what the neurons are supposed to do at each timestep.
from PymoNNto import *
net = Network()
NeuronGroup(net=net, tag='my_neurons', size=100, behavior={})
SynapseGroup(net=net, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
net.initialize()
net.simulate_iterations(1000)Each Behavior Module has the following layout where initialize is called when the Network is initialized, while
iteration is called repeatedly every timestep. neurons points to the parent neuron group the behavior belongs to.
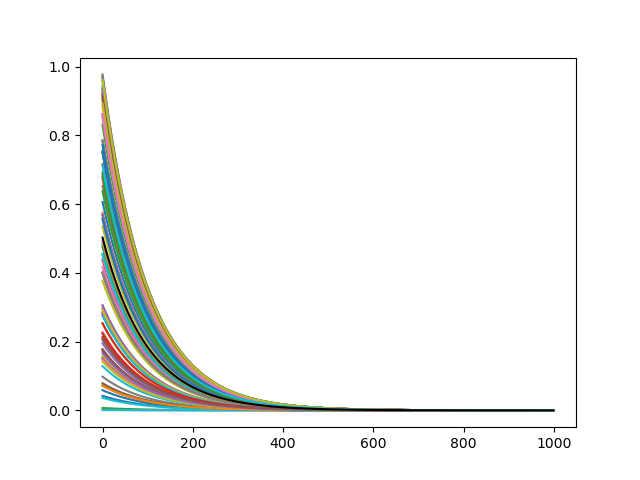
In this example we define a variable activity and a decay_factor. The activity-vector is initialized with random values for each neuron. At each timestep the activity-vector is decreased.
class Basic_Behavior(Behavior):
def initialize(self, neurons):
neurons.activity = neurons.vector('uniform')
self.decay_factor = 0.99
def iteration(self, neurons):
neurons.activity *= self.decay_factorWhen we combine the previous code blocks we can add the Basic_Behavior to the NeuronGroup.
To plot the neurons activity over time, we also have to create a Recorder. Here the activity and the mean-activity are stored at each timestep.
At the end the data is plotted.
PymoNNto has a tagging system to make access to the NeuronGroups, SynapseGroups, Behaviors and recorded variables inside of the network more convenient.
The number in front of each behavior (1 and 9) have to be positive and determine the order of execution for each module inside and accross NeuronGroups.
from PymoNNto import *
class Basic_Behavior(Behavior):
def initialize(self, neurons):
neurons.activity = neurons.vector('uniform')
self.decay_factor = 0.99
def iteration(self, neurons):
neurons.activity *= self.decay_factor
net = Network()
NeuronGroup(net=net, tag='my_neurons', size=100, behavior={
1: Basic_Behavior(),
9: Recorder(['activity', 'np.mean(activity)'], tag='my_recorder')
})
SynapseGroup(net=My_Network, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
net.initialize()
net.simulate_iterations(1000)
import matplotlib.pyplot as plt
plt.plot(net['activity', 0])
plt.plot(net['np.mean(activity)', 0], color='black')
plt.show()To access the tagged objects we can use the [] operator. ['my_tag'] gives you a list of all objects tagged with my_tag. Here are some examples:
My_Network = net
My_Neurons = net['my_neurons']
=> [<PymoNNto.NetworkCore.Neuron_Group.NeuronGroup object at 0x00000195F4878670>]
My_Network['my_recorder']
My_Neurons['my_recorder']
=> [<PymoNNto.NetworkBehavior.Recorder.Recorder.Recorder object at 0x0000021F1B61D5E0>]
My_Neurons['activity']
My_Neurons['my_recorder', 0]['activity']
=> [[array(data iteration 1), array(data iteration 2), array(data iteration 3), ...]]
My_Neurons['activity', 0]
is equivalent to
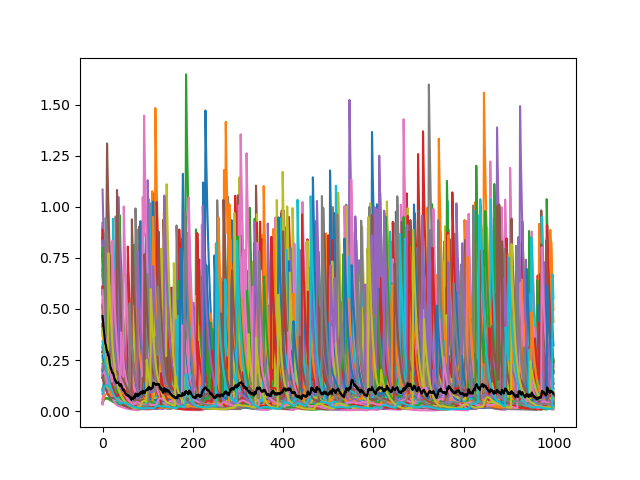
My_Neurons['activity'][0] We can add more behavior modues to make the activity of the neurons more complex. Here the module Input_Behavior is added. In initialize the synapse matrix is created, which stores one weight-value from each neuron to each neuron. The Function iteration defines how the information is propagated to each neuron (dot product) and adds some term for random input.
The for loops are not neccessary here, because we only have one SynapseGroup. This solution, however, also works with multiple Neuron- and SynapseGroups. With synapse.src and synapse.dst you can access the source and destination NeuronGroups assigned to a SynapseGroup.
from PymoNNto import *
class Input_Behavior(Behavior):
def initialize(self, neurons):
for synapse in neurons.synapses(afferent,'GLUTAMATE'):
synapse.W = synapse.matrix('uniform', density=0.1)
def iteration(self, neurons):
for synapse in neurons.synapses(afferent,'GLUTAMATE'):
neurons.activity += synapse.W.dot(synapse.src.activity) / synapse.src.size
neurons.activity += neurons.vector('uniform', density=0.01)
class Basic_Behavior(Behavior):
def initialize(self, neurons):
neurons.activity = neurons.vector('uniform')
self.decay_factor = 0.99
def iteration(self, neurons):
neurons.activity *= self.decay_factor
net = Network()
NeuronGroup(net=net, tag='my_neurons', size=100, behavior={
1: Basic_Behavior(),
2: Input_Behavior(),
9: Recorder(['activity', 'np.mean(activity)'], tag='my_recorder')
})
SynapseGroup(net=net, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
net.initialize()
net.simulate_iterations(1000)
import matplotlib.pyplot as plt
plt.plot(My_Network['activity', 0])
plt.plot(My_Network['np.mean(activity)', 0], color='black')
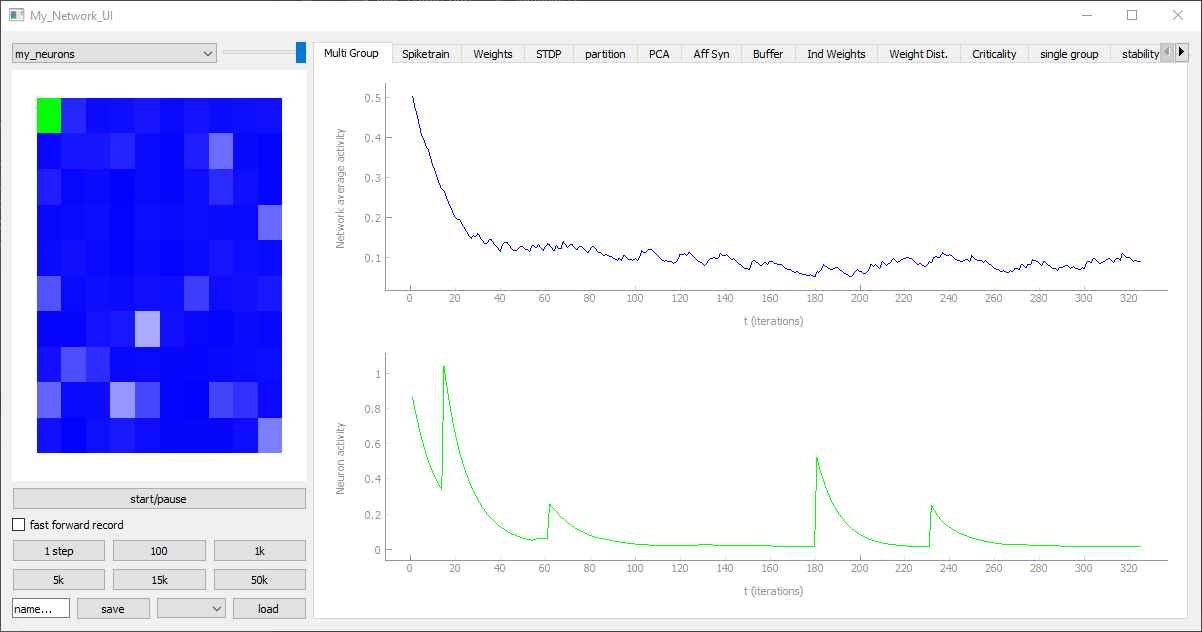
plt.show()If we want to controll and evaluate our model in realtime we can replace the pyplot functions, the recorder and the simulate_iterations with the following Code lines. Similar to the NeuronGroup, the Network_UI is also modular and consists of multiple UI_modules. We can choose them ourselfes or, like in this case, take some default_modules, which should work with most networks. One addition we have to make is to give NeuronGroup some shape with the help of the get_squared_dim(100) function, which returns a 10x10 grid NeuronDimension object. The neuron-behaviors are not affected by this. The NeuronGroup only receives some additional values like width, height, depth and the vectors x, y, z. The neurons positions can be plotted with plt.scatter(My_Neurons.x, My_Neurons.y)
from PymoNNto import *
class Input_Behavior(Behavior):
def initialize(self, neurons):
for synapse in neurons.synapses(afferent,'GLUTAMATE'):
synapse.W = synapse.matrix('uniform', density=0.1)
def iteration(self, neurons):
for synapse in neurons.synapses(afferent,'GLUTAMATE'):
neurons.activity += synapse.W.dot(synapse.src.activity) / synapse.src.size
neurons.activity += neurons.vector('uniform', density=0.01)
class Basic_Behavior(Behavior):
def initialize(self, neurons):
neurons.activity = neurons.vector('uniform')
self.decay_factor = 0.99
def iteration(self, neurons):
neurons.activity *= self.decay_factor
net = Network()
NeuronGroup(net=net, tag='my_neurons', size=get_squared_dim(100), behavior={
1: Basic_Behavior(),
2: Input_Behavior(),
9: Recorder(['activity', 'np.mean(activity)'], tag='my_recorder')
})
SynapseGroup(net=net, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
net.initialize()
# My_Network.simulate_iterations(1000)
from PymoNNto.Exploration.Network_UI import *
my_UI_modules = get_default_UI_modules(['activity'], ['W'])
Network_UI(net, modules=my_UI_modules, label='My_Network_UI', group_display_count=1).show()