-
-
Notifications
You must be signed in to change notification settings - Fork 2.2k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Phase cross correlation : uncorrect shift in some conditions #5460
Comments
|
The recently opened #5459 indicated that there is a bug in the function. |
|
Indeed, it seems making the fix suggested in #5459 results in the expected result for both cases here |
|
Hi, Indeed, now the test case above returns good results (the same for my more complicated one :) ) BEFORE introducing the bugfix : AFTER introducing the bugfix : Note that when commenting the gausssian filter operation, the shift recovers a correct value. |
|
I could not reproduce the failure in your example when using the branch corresponding to #5461. Can you maybe try the following variant, and see if it helps? (It adds a multiple of eps only to extremely small values, but doesn't modify the rest) image_product = src_freq * target_freq.conj()
eps = np.finfo(image_product.real.dtype).eps
image_product /= np.maximum(np.abs(image_product), 100 * eps) |
|
hmmm .. embarrassing. What could be wrong ? |
|
I have performed test with different seed. |
|
I am experiencing the same thing, depending on the seed the phase correlation can either fail or succeed whereas the cross correlation is always good in this case. Now, this type of image is really a texture where most of the information is contained in the frequencies amplitude. If you remove the gaussian filter the phase correlation does not fail because on the blobs edges all the frequencies are in phase. In my opinion it is the user responsibility to use a registration algorithm accordingly to a given problem, in this case the function is named phase cross correlation and it should do so or maybe it should be renamed (with an optional argument to return the phase correlation?). |
|
I agree with you @JulesScholler |


Hello,
Here is a simple case that illustrates problems that I meet in a more complicated one.
In some "conditions" (?), the calculated shift is not correct.
In the pseudo 1D example below, I play with 2 parameters:
length : which is the length of my trench of my 'profile'
shift : which is the shift applied between the 2 'profiles'
with length=20, the calculated shifts are correct in the both cases
with length=10, the calculated shift is not correct in the case of shift=20.
length = 10 / shift = 15 : OK
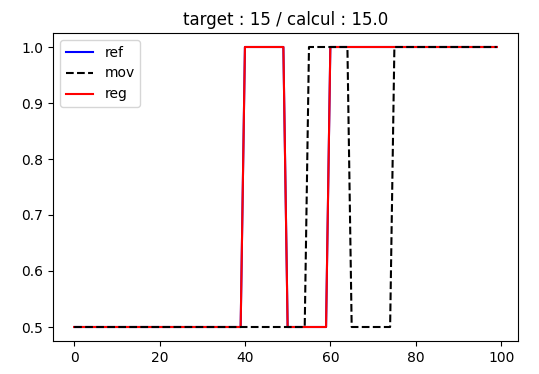
length = 10 / shift = 20 : NOK

What the reason of this ?
Thanks
Patrick
The text was updated successfully, but these errors were encountered: