-
-
Notifications
You must be signed in to change notification settings - Fork 2.2k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
peak_local_max attribute 'min_distance' issue #5598
Comments
|
Thanks for opening this issue @taroko-mooncake! Could you please provide us a full, runnable example for us to check this bug? |
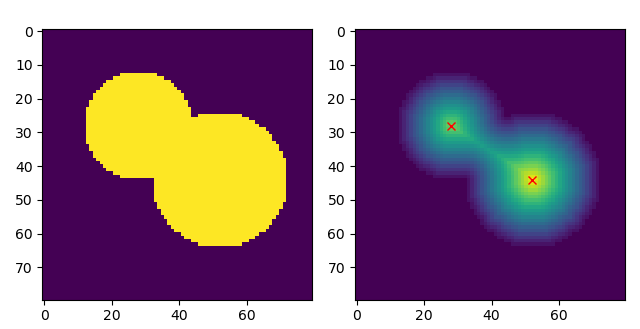
# Generate an initial image with two overlapping circles
x, y = np.indices((80, 80))
x1, y1, x2, y2 = 28, 28, 44, 52
r1, r2 = 16, 20
mask_circle1 = (x - x1)**2 + (y - y1)**2 < r1**2
mask_circle2 = (x - x2)**2 + (y - y2)**2 < r2**2
image = np.logical_or(mask_circle1, mask_circle2)
# Now we want to separate the two objects in image
# Generate the markers as local maxima of the distance to the background
distance = ndi.distance_transform_edt(image)
coords = peak_local_max(distance, footprint=np.ones((3, 3)), labels=image)` |
|
@taroko-mooncake, can you please be more precise in your bug report. Displaying the result of your provided example gives fig, (ax0, ax1) = plt.subplots(1, 2)
ax1.imshow(distance)
ax0.imshow(image)
ax1.plot(coords[:, 1], coords[:, 0], 'rx') |
|
Yes the algorithm works but if you apply 'min_distance' attribute, nothing changes. # Generate an initial image with two overlapping circles
x, y = np.indices((80, 80))
x1, y1, x2, y2 = 28, 28, 44, 52
r1, r2 = 16, 20
mask_circle1 = (x - x1)**2 + (y - y1)**2 < r1**2
mask_circle2 = (x - x2)**2 + (y - y2)**2 < r2**2
image = np.logical_or(mask_circle1, mask_circle2)
# Now we want to separate the two objects in image
# Generate the markers as local maxima of the distance to the background
distance = ndi.distance_transform_edt(image)
coords = peak_local_max(distance, footprint=np.ones((3, 3)), labels=image, min_distance=70)`There is something about min_distance that is causing a bug but I can't tell you anymore than that. |
|
@taroko-mooncake, as the Note section stipulates in the doc:
So the peak candidates are selected from this image obtained using import numpy as np
from scipy import ndimage as ndi
import matplotlib.pyplot as plt
from skimage.feature import peak_local_max
# Generate an initial image with two overlapping circles
x, y = np.indices((80, 80))
x1, y1, x2, y2 = 28, 28, 44, 52
r1, r2 = 16, 20
mask_circle1 = (x - x1)**2 + (y - y1)**2 < r1**2
mask_circle2 = (x - x2)**2 + (y - y2)**2 < r2**2
image = np.logical_or(mask_circle1, mask_circle2)
# Now we want to separate the two objects in image
# Generate the markers as local maxima of the distance to the background
distance = ndi.distance_transform_edt(image)
footprint = np.ones((3, 3))
# Show peak candidates
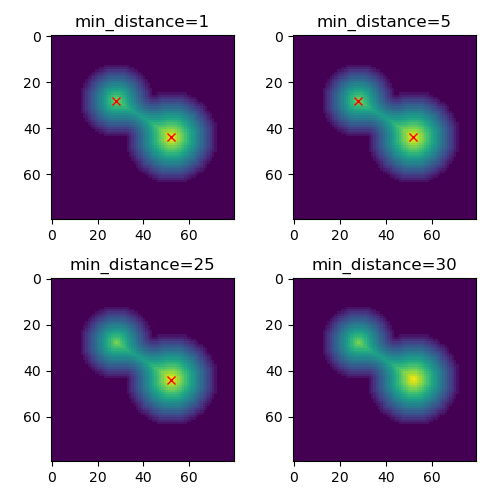
plt.imshow(ndi.maximum_filter(distance, footprint=footprint,
mode='constant') == distance)Now with different values for # Show the output with different min_distance
fig, ax_list = plt.subplots(2, 2, figsize=(5, 5))
for ax, min_dist in zip(ax_list.ravel(), [1, 5, 25, 30]):
coords = peak_local_max(distance, footprint=np.ones((3, 3)),
min_distance=min_dist)
ax.imshow(distance)
ax.plot(coords[:, 1], coords[:, 0], 'rx')
ax.set_title(f"min_distance={min_dist}")
fig.tight_layout()
plt.show()Please note that with |
|
Which version of |
|
BTW, If you don't mind, I will convert this issue to a Discussion. |
comment moved back to issues from Discussion
|

Description
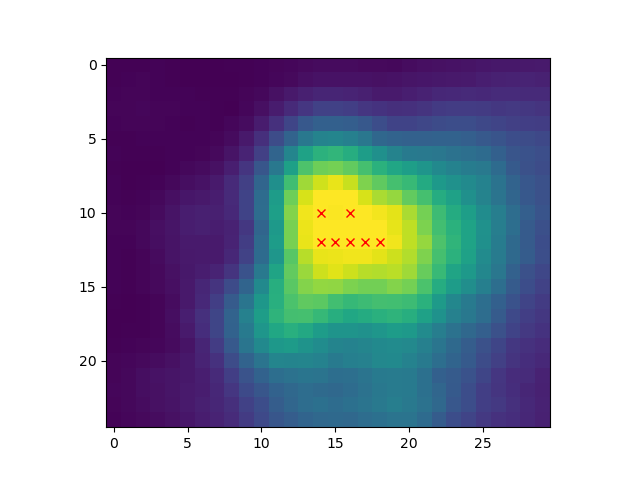
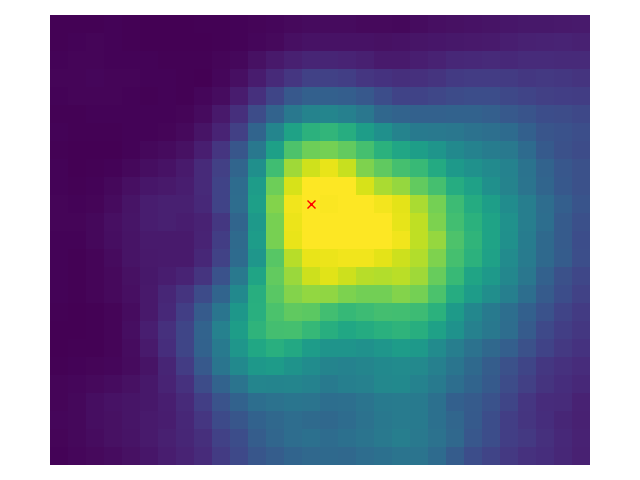
The attribute 'min_distance' in peak_local_max function is not working. Distance can be set at any integer and coordinates of centres remain unchanged.
Way to reproduce
Version information
The text was updated successfully, but these errors were encountered: