Enhance gallery example on color separation of stained tissues #6333
Labels
📄 type: Documentation
Updates, fixes and additions to documentation
😴 Dormant
no recent activity
💬 Discussion
Moved from now deleted Discussion https://github.com/scikit-image/scikit-image/discussions/6065
Originally posted by mkcor November 26, 2021
Hello,
I'm starting this discussion as per #5172 (comment).
First, let me thank @alessiamarcolini!
Wonderful, you've definitely upped my search game! 💪
@alexdesiqueira I was thinking of enhancing the current example with an additional section based on an additional image (i.e., an H&E image such as the one @alessiamarcolini spotted).
I think the three subplots for the IHC image should be: Original, H, DAB; for the H&E image, they should be: Original, H, E (as the name suggests). The H&E sum does not make sense in the IHC case, because no Eosin staining was used in the first place... (I'm trying to tell a story which relates to real use cases in the life sciences, I'm not trying to rearrange subplots so that they render nicely...)
But...
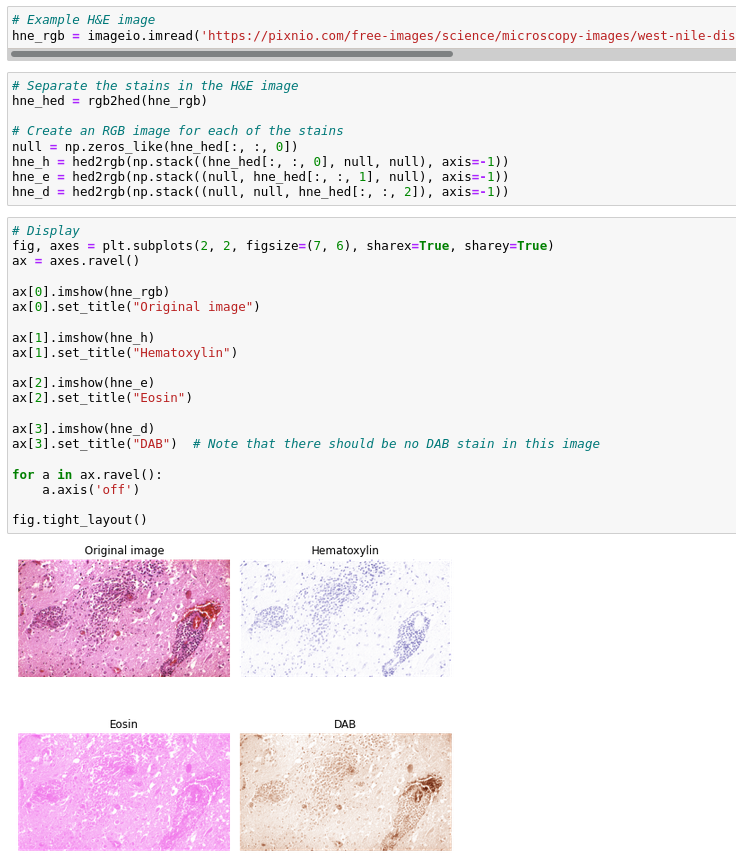
this H&E image has all three stains!! Hematoxylin in blue-purple, Eosin in pink, and DAB in brown. How come? @crisluengo I expected no DAB stain. 😕
...of course you could create a clean image to unmix by setting
hne_hed[:,:,2]=0, and thenhne_rgb=hed2rgb(hne_hed).Looking through the documentation, I see there is a matrix for Hematoxylin + DAB (
rgb_from_hdx), but no Hematoxylin + Eosin; instead there is Hematoxylin + Eosin + DAB (rgb_from_hed). This latter is the only one with a function made around it, note that all this does isseparate_stains(rgb, hed_from_rgb), it's just as easy to callseparate_stains(rgb, hdx_from_rgb). And it'd be just as easy to create a matrix for Hematoxylin + Eosin. If you do that, this whole demo will be cleaner and easier to explain.I also see that the
rgb_from_hdxmatrix is a 3x3 matrix, meaning it outputs 3 channels. I presume the 3rd channel is a bogus channel. Why not make this a 3x2 matrix? Is there a reason skimage always wants 3 channels?The Hema+DAB array gets a 3rd row composed as the cross product, which is a color orthogonal to the other two. This should make the inverse robust. But it is also possible to do a pseudo-inverse of the 2x3 matrix, yielding a 3x2 matrix that is equal to the first two columns of the
hdx_from_rgbconstructed above. Avoiding that additional channel would be neat, because then there is no confusion as to what that channel means.I also see that
rgb_from_bex(Methyl Blue + Eosin) uses a different color for Eosin ([0.07, 0.99, 0.11]for HED,[0.092789, 0.954111, 0.283111]for BEX). This must have been someone else in another lab measuring an Eosin stain on one other slide... :)Oh, I forgot that we already have
data.skin()... 🤦♀️https://scikit-image.org/docs/dev/auto_examples/data/plot_scientific.html
crisluengo
on Mar 2
Here's a suggestion for how to modify skimage's stain unmixing functionality. HED is just confusing and useless. If there are two stains on the slide, you want the stain unmixing to yield two channels, not three.
crisluengo@431aa9a
These changes involve backward-incompatible changes, so I'm not making this into a PR. I just made those changes to help a conversation about this topic.
grlee77
on Mar 2
Yeah, I think that could be helpful. We are actually planning on making an
skimage2release later this year with a limited number of breaking changes so that would be a good opportunity to do this. If you want to open PR I can tag it for that purpose. (Mainly, I just don't want this to get lost as we are currently in the process of migrating GitHub Discussions back to the issues page).On a related note in the RAPIDS cuCIM package, the NVIDIA team proposed a PR for a different stain normalization than the one in
skimage. That difference is why it isn't currently under thecucim.skimagenamespace, but I was planning to review and see if we should port it here. I haven't reviewed it properly yet, but on initial glance it does look like it produces a 2-channel H & E image.crisluengo
on Mar 3
@grlee77 See #6274.
There are two options there, the commit I referenced earlier, and an even more invasive one I just did now, after hearing about skimage2. This new version, I hope, makes the stain unmixing look and work differently from color space conversions, to avoid the confusion I've seen so far.
The text was updated successfully, but these errors were encountered: