New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
PCA on sparse, noncentered data #12794
Comments
|
Did you see the TruncatedSVD estimator? In the current implementation, it should be equivalent to PCA on centered data, except that it does not necessarily center the data and also supports sparse input. |
|
Yes, I am aware of TruncatedSVD, but that's different. It's true that SVD on centered data is PCA, but SVD on noncentered data is not equivalent to PCA. It's easy to see the difference with a simple example: >>> x = datasets.load_boston().data
>>> x.mean(axis=0) # Not centered!
array([ 3.61352356, 11.36363636, 11.13677866, 0.06916996,
0.55469506, 6.28463439, 68.57490119, 3.79504269,
9.54940711, 408.23715415, 18.4555336 , 356.67403162,
12.65306324])
>>> sksvd = decomposition.TruncatedSVD(n_components=6)
>>> sksvd.fit(x)
>>> sksvd.singular_values_
array([12585.18158999, 3445.97405949, 645.75710949, 402.05046109,
158.96461248, 121.50293599])
>>> skpca = decomposition.PCA(n_components=6)
>>> skpca.fit(x)
>>> skpca.singular_values_
array([3949.60823205, 1776.63071036, 642.86374839, 366.98207453,
158.634553 , 118.64982369])Now, I'm dealing with huge sparse matrices. If I densify them, they would not fit into memory, so centering the matrix is not an option (we lose all sparsity with centering). Yes, I could perform incremental PCA, loading chunks of the matrix into memory at a time, but that's really slow. Yes, I could perform SVD on these matrices, which is fine, but not the same as PCA. Now, like I said in my first comment, this can be done i.e. we can compute PCA on noncentered data directly, without ever having to directly perform centering. This is what I'm missing. I hope I've it's clear enough this time. PCA is possible and is very fast on sparse matrices, and I feel it really should be included in scikit-learn. |
|
It sounds like we should certainly consider a pull request
|
I am confused. It seems that you want PCA with non centered data which is as you mentioned, different from PCA. So it seems that you want the truncated SVD, isn't it? |
|
Perhaps I've been unclear. I'll try to be more clear this time :) What we know So this is nothing new. We typically compute PCA via the SVD. And when X is centered, SVD is the same as PCA. Otherwise they are different. The problem We can still do SVD, but since X is not centered, this is not the same as PCA. PCA is often nicer because it's easier to interpret things. It's pretty hard to interpret an SVD (or I just might not be aware of it). The solution To achieve this, we take the already implemented randomized SVD algorithm, and add a couple of negative terms, which account for centering. The changes to the algorithm are very minor. The randomized SVD implemented here is great and could be extended with just a couple of terms, and we could have it compute both the SVD and PCA. Setting a single flag e.g. Conclusion I could work on this if needed, but I am not at all familliar with the codebase, and have fairly limited time. |
|
Oh ok got it. Make sense then to have a PR.
|
|
Yeah I think it's clear and I think we probably want it but we need to research the math and check speed tradeoffs and complexity of adding the method etc. |
|
Actually, I've just briefly looked at the implementations and it seems to be astonishingly simple. Randomized methods use power iteration to increase the spectral gap, which is just a fancy way of saying multiplication by the original matrix X multiple times. SVD already does this in the current implementation. I don't think I've explained this properly (clearly I need to work on my explanations), but it's just a clever application of the distributive property of the dot product. |
|
If I am not wrong (I never really read the code before) it looks like the solver which is the one used by default in the |
|
So, I've played around with this a little bit and I think these are all the changes needed int the randomized SVD solver. The only real change is that whenever we apply a non-centered matrix A to any other matrix, we distribute the mean subtraction like so: (A - m) X = AX - mX, avoiding the need to ever center A. diff --git a/sklearn/utils/extmath.py b/sklearn/utils/extmath.py
index 19df5b161..0ab75721e 100644
--- a/sklearn/utils/extmath.py
+++ b/sklearn/utils/extmath.py
@@ -147,6 +147,7 @@ def safe_sparse_dot(a, b, dense_output=False):
def randomized_range_finder(A, size, n_iter,
power_iteration_normalizer='auto',
+ subtract_mean=False,
random_state=None):
"""Computes an orthonormal matrix whose range approximates the range of A.
@@ -211,28 +212,39 @@ def randomized_range_finder(A, size, n_iter,
else:
power_iteration_normalizer = 'LU'
+ if subtract_mean:
+ c = np.mean(A, axis=0).reshape((1, -1))
+ applyA = lambda X: safe_sparse_dot(A, X) - safe_sparse_dot(c, X)
+ applyAT = lambda X: safe_sparse_dot(A.T, X) - \
+ safe_sparse_dot(c.T, Q.sum(axis=0).reshape((1, -1)))
+ else:
+ applyA = lambda X: safe_sparse_dot(A, X)
+ applyAT = lambda X: safe_sparse_dot(A.T, X)
+
+ Q = applyA(Q)
+
# Perform power iterations with Q to further 'imprint' the top
# singular vectors of A in Q
for i in range(n_iter):
if power_iteration_normalizer == 'none':
- Q = safe_sparse_dot(A, Q)
- Q = safe_sparse_dot(A.T, Q)
+ Q = applyAT(Q)
+ Q = applyA(Q)
elif power_iteration_normalizer == 'LU':
- Q, _ = linalg.lu(safe_sparse_dot(A, Q), permute_l=True)
- Q, _ = linalg.lu(safe_sparse_dot(A.T, Q), permute_l=True)
+ Q, _ = linalg.lu(applyAT(Q), permute_l=True)
+ Q, _ = linalg.lu(applyA(Q), permute_l=True)
elif power_iteration_normalizer == 'QR':
- Q, _ = linalg.qr(safe_sparse_dot(A, Q), mode='economic')
- Q, _ = linalg.qr(safe_sparse_dot(A.T, Q), mode='economic')
+ Q, _ = linalg.qr(applyAT(Q), mode='economic')
+ Q, _ = linalg.qr(applyA(Q), mode='economic')
# Sample the range of A using by linear projection of Q
# Extract an orthonormal basis
- Q, _ = linalg.qr(safe_sparse_dot(A, Q), mode='economic')
+ Q, _ = linalg.qr(Q, mode='economic')
return Q
def randomized_svd(M, n_components, n_oversamples=10, n_iter='auto',
power_iteration_normalizer='auto', transpose='auto',
- flip_sign=True, random_state=0):
+ flip_sign=True, subtract_mean=False, random_state=0):
"""Computes a truncated randomized SVD
Parameters
@@ -333,11 +345,20 @@ def randomized_svd(M, n_components, n_oversamples=10, n_iter='auto',
# this implementation is a bit faster with smaller shape[1]
M = M.T
- Q = randomized_range_finder(M, n_random, n_iter,
- power_iteration_normalizer, random_state)
+ Q = randomized_range_finder(
+ M,
+ size=n_random,
+ n_iter=n_iter,
+ power_iteration_normalizer=power_iteration_normalizer,
+ subtract_mean=subtract_mean,
+ random_state=random_state,
+ )
# project M to the (k + p) dimensional space using the basis vectors
B = safe_sparse_dot(Q.T, M)
+ if subtract_mean:
+ c = np.mean(M, axis=0).reshape((1, -1))
+ B -= np.dot(c.T, Q.sum(axis=0).reshape((1, -1)))
# compute the SVD on the thin matrix: (k + p) wide
Uhat, s, V = linalg.svd(B, full_matrices=False)I've run all the tests for TruncatedSVD, and they pass. In the PCA learner, there's one problematic line: total_var = np.var(X, ddof=1, axis=0)which fails on sparse matrices. I'd expect something like sparse variance to be already implemented, but I'm not familliar with the codebase and don't know where to look. If not, it should be relatively straightforward to implement this as well. |
|
For the variance I guess you can just use the formula: E[X^2] - (E[X])^2 |
in sklearn you have |
|
I looked into the paper and they actually only speak of the SVD which explain why our implementation does not remove the mean. Then, your proposal looks good to me. Since that we already have @pavlin-policar thanks a lot for the clarifications and the patience ;) |
|
@pavlin-policar would you be keen on opening a PR with the changes? |
|
Yeah, sure. I'll give it a shot. @jeremiedbb Thank's that's going to make things easier. |
|
I've opened #12841 which implements the changes I described above. |
|
#12319 would be a (faster) alternative to 'randomized' for sparse matrices that also allows running matrix-free SVD, i.e. on a matrix, given by a function. In particular, performing implicit data centering, without changing the data. For that matter, the ARPACK SVD solver is also matrix-free by design. Thus, one should really make the implicit data centering to be solver agnostic, allowing any matrix-free SVD solver to be used, rather than concentrating just on 'randomized' |
|
@lobpcg -- I'm really interested in allowing implicit data centering using your solver. I checked out the |
@atarashansky If you want to code specifically LOBPCG for PCA with implicit data centering and submit as a PR to scikit, you probably want to pick up #12841, update it to resolve the conflicts, and add the support for If you want to run a code specifically with LOBPCG for PCA with implicit data centering for yourself, it is now easy, since PCA is just SVD(X - means), and LOBPCG for SVD is already merged into scipy in scipy/scipy#10830 You just need to code the LinearOperator A multiplying (X - means) * Q as X*Q - means * Q and similar for the transpose to pass to svds in https://github.com/scipy/scipy/blob/0db79f6a56ba8187380288372993800331e9f9ba/scipy/sparse/linalg/eigen/arpack/arpack.py since now: |
|
@lobpcg -- I have a pretty naive question: The first dimension of Q will be the minimum dimension of the input data, right? If I have more features (m) than samples (n), then Q will be of shape (n x k). But 'means' will be shape (1 x m). So, in " X*Q - means * Q", is 'means' the mean of either columns or rows depending on the shape of Q? And if its the mean of the rows, is that still equivalent to PCA (SVD on feature-centered data)? EDIT: Ah, nevermind, I just got confused by the notation. I got it working! Thanks so much! |
|
For others who are looking for a solution while waiting for the aforementioned PRs to go through, here is what worked for me: Anecdotally, on my dataset (120,000 x 10,000), |
|
@atarashansky |
|
Does order of operations mitigate this issue? I was able to do what you suggest for the If X is (n x m) and mu is (1 x m), then mean centering is X * x - 1 * mu * x where 1 * mu is an outer product between a column of 1s of size n and the means. In the code, however, we don't need the column of ones because mu * x will be broadcasted and subtracted from every row in X * x. I ran into confusion when I tried doing the same for the transpose, (X^T - mu^T * 1^T) * x = X^T * x - mu^T * 1^T * x, as now the ones are in the middle of the product.. |
|
@atarashansky I actually misread your code yesterday and made technically wrong comments (now deleted), but you still were able to see my concern. May be try efficiently implementing mu^T ( 1^T * x) instead of (mu^T * 1^T ) x, also keeping in mind that x is a matrix in rmatmat. May be something like ( x^T *1)^T could work? You can try using a similar trick to code the X-part of rmatvec and rmatmat without explicitly forming X.T.conj() (which may double the memory use, also storing X^T ?) using instead something like (x^T*X)^T. In https://www.mathworks.com/matlabcentral/fileexchange/48-locally-optimal-block-preconditioned-conjugate-gradient, I feed LOBPCG the function funA = @(v)((X*v)'*X)') for A = X'*X to perform svd(X) Please let me know if you find the "optimal" way to code it, i.e., without doing anything to X and creating no new matrices of the size larger than size(x). Another fun option to try is to actually generate (some of or all) 4 matrices resulting from the product (X^T - mu^T * 1^T) *(X - 1 * mu). Depending on the size and sparsity of X, as well as on the convergence speed of LOBPCG, that might be a competing option. I have found testing svd(X) with LOBPCG, that explicitly computing X'X once might be beneficial speed-wise compared to the no-extra-memory way feeding funA = @(v)((Xv)'*X)') if size(X) is moderate. |
|
I tried running the function with and without the mean centering on a sparse 30,000 x 30,000 dataset, and the memory usage and timings are identical according to I'm sure the way I coded it is indeed not optimal, but it seems that, practically, it makes no difference, at least not in the way |
|
Thanks for checking! 'arpack' is pre-compiled - I do not know if memit accurately captures the memory usage in such a case. |
|
@atarashansky This is an amazing code snippet! I haven't tried it out myself but assuming it works as advertised, this is really cool. I wasn't aware that one can feed @pavlin-policar You have that old PR that implements PCA for sparse matrix with the randomized solver. This looks like a great way to implement the same using |
|
@dkobak -- To answer your question about how the interfacing with ARPACK works in this case, from @lobpcg above:
Apparently, the ARPACK solver is also matrix-free by design. In ... And then in The ARPACK class just accepts the dot product functions as input, meaning the Hermetian and input data matrix never need to be explicitly generated. |
It doesn't. All svds sparse algorithms in SciPy are matrix-free, i.e., they don't modify the data matrix M in any way, so the matrix access is read only to perform products by M. Moreover, the matrix can be accessed only via a function that performs the product, e.g., the matrix entries could even be computed on the fly as needed. |
|
Got it, thank you. |
|
I have randomized SVD now working along with ARPACK, though the code is still at the proof of concept stage. Apparently As a workaround I used I tried to debug using a debug build of Python but I've only figured out that the object corresponding to the The code is in #24415. |
|
@andportnoy Yes, I am sure that LinearOperator must be on the LHS. Your workaround looks reasonable. If you start using LOBPCG in addition to ARPACK, make sure to specify also matmat, not just matvec, if the definition of the LinearOperator Please file a bug report if you believe that this is a bug: |
|
Got it, thanks. Going to work on @lobpcg next (and PROPACK as well). I'll spend some more time trying to root cause the error (why isn't this struct field populated? is that expected? can it be fixed?). Will file a bug report in SciPy. |
|
I've looked at the error, The easiest way to reproduce is NumPy very aggressively converts all kinds of objects to arrays, which can be useful but in this case it's slightly confusing. I would have preferred an error message along the lines of "cannot coerce a LinearOperator to a NumPy array". Relevant stack traces: max_ndim set to 0#0 update_shape (curr_ndim=0, max_ndim=0x7fffffffc3b0, out_shape=0x7fffffffc580, new_ndim=0,
new_shape=0x0, sequence=0 '\000', flags=0x7fffffffc494)
at numpy/core/src/multiarray/array_coercion.c:572
#1 0x00007fffe98ec220 in handle_scalar (
obj=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, curr_dims=0, max_dims=0x7fffffffc3b0, out_descr=0x7fffffffc530,
out_shape=0x7fffffffc580, fixed_DType=0x0, flags=0x7fffffffc494, DType=0x0)
at numpy/core/src/multiarray/array_coercion.c:738
#2 0x00007fffe98ed073 in PyArray_DiscoverDTypeAndShape_Recursive (
obj=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, curr_dims=0, max_dims=32, out_descr=0x7fffffffc530,
out_shape=0x7fffffffc580, coercion_cache_tail_ptr=0x7fffffffc470, fixed_DType=0x0,
flags=0x7fffffffc494, never_copy=0) at numpy/core/src/multiarray/array_coercion.c:1133
#3 0x00007fffe98ed57e in PyArray_DiscoverDTypeAndShape (
obj=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, max_dims=32, out_shape=0x7fffffffc580, coercion_cache=0x7fffffffc538,
fixed_DType=0x0, requested_descr=0x0, out_descr=0x7fffffffc530, never_copy=0)
at numpy/core/src/multiarray/array_coercion.c:1289
#4 0x00007fffe9913346 in PyArray_FromAny (
op=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, newtype=0x0, min_depth=0, max_depth=0, flags=0, context=0x0)
at numpy/core/src/multiarray/ctors.c:1629
#5 0x00007fffe9ae0a92 in convert_ufunc_arguments (ufunc=0x7fffe6c58c00, full_args=...,
out_op=0x7fffffffc9b0, out_op_DTypes=0x7fffffffcab0, force_legacy_promotion=0x7fffffffc7cb "",
allow_legacy_promotion=0x7fffffffc7cc "\001", promoting_pyscalars=0x7fffffffc7cd "",
order_obj=0x0, out_order=0x7fffffffc7d0, casting_obj=0x0, out_casting=0x7fffffffc7d4,
subok_obj=0x0, out_subok=0x7fffffffc7ca "\001", where_obj=0x0, out_wheremask=0x7fffffffc7f8,
keepdims_obj=0x0, out_keepdims=0x7fffffffc7d8) at numpy/core/src/umath/ufunc_object.c:977
#6 0x00007fffe9aea9ed in ufunc_generic_fastcall (ufunc=0x7fffe6c58c00, args=0x7fffffffcef0,
len_args=2, kwnames=0x0, outer=0 '\000') at numpy/core/src/umath/ufunc_object.c:4896
#7 0x00007fffe9aeae8e in ufunc_generic_vectorcall (ufunc=<numpy.ufunc at remote 0x7fffe6c58c00>,
args=0x7fffffffcef0, len_args=2, kwnames=0x0) at numpy/core/src/umath/ufunc_object.c:5011
#8 0x00007ffff7b7a1cc in _PyObject_VectorcallTstate (tstate=0x555555577360,
callable=<numpy.ufunc at remote 0x7fffe6c58c00>, args=0x7fffffffcef0, nargsf=2, kwnames=0x0)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Include/cpython/abstract.h:114
#9 0x00007ffff7b7c5c8 in object_vacall (tstate=0x555555577360, base=0x0,
callable=<numpy.ufunc at remote 0x7fffe6c58c00>, vargs=0x7fffffffcf50)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Objects/call.c:734
#10 0x00007ffff7b7cba4 in PyObject_CallFunctionObjArgs (
callable=<numpy.ufunc at remote 0x7fffe6c58c00>)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Objects/call.c:841
#11 0x00007fffe9a1c435 in PyArray_GenericBinaryFunction (
m1=<numpy.ndarray at remote 0x7fff95382ec0>,
m2=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, op=<numpy.ufunc at remote 0x7fffe6c58c00>)
at numpy/core/src/multiarray/number.c:270
#12 0x00007fffe9a1c934 in array_matrix_multiply (m1=<numpy.ndarray at remote 0x7fff95382ec0>,
m2=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>) at numpy/core/src/multiarray/number.c:347
#13 0x00007ffff7b4bb1f in binary_op1 (v=<numpy.ndarray at remote 0x7fff95382ec0>,
w=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, op_slot=272, op_name=0x7ffff7e3877e "@")
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Objects/abstract.c:891
#14 0x00007ffff7b4bcd8 in binary_op (v=<numpy.ndarray at remote 0x7fff95382ec0>,
w=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>, op_slot=272, op_name=0x7ffff7e3877e "@")
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Objects/abstract.c:930
#15 0x00007ffff7b4c65a in PyNumber_MatrixMultiply (v=<numpy.ndarray at remote 0x7fff95382ec0>,
w=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7fffea083b60>) at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Objects/abstract.c:1128
#16 0x00007ffff7cea99b in _PyEval_EvalFrameDefault (tstate=0x555555577360,
f=Frame 0x5555555d8f50, for file /home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py, line 19, in <module> (), throwflag=0)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/ceval.c:2015
#17 0x00007ffff7ce559f in _PyEval_EvalFrame (tstate=0x555555577360,
f=Frame 0x5555555d8f50, for file /home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py, line 19, in <module> (), throwflag=0)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Include/internal/pycore_ceval.h:46
#18 0x00007ffff7cfdd52 in _PyEval_Vector (tstate=0x555555577360, con=0x7fffffffe010,
locals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated), args=0x0, argcount=0, kwnames=0x0)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/ceval.c:5065
#19 0x00007ffff7ce86ee in PyEval_EvalCode (co=<code at remote 0x7fffea043780>,
globals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated),
locals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated))
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/ceval.c:1134
#20 0x00007ffff7d69719 in run_eval_code_obj (tstate=0x555555577360, co=0x7fffea043780,
globals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated),
locals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated))
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/pythonrun.c:1291
#21 0x00007ffff7d6981b in run_mod (mod=0x5555555fd440,
filename='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py',
globals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated),
locals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated), flags=0x7fffffffe268, arena=0x7fffea0726e0)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/pythonrun.c:1312
#22 0x00007ffff7d694ad in pyrun_file (fp=0x555555588cf0,
filename='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', start=257,
globals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated),
locals={'__name__': '__main__', '__doc__': None, '__package__': None, '__loader__': <SourceFileLoader(name='__main__', path='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py') at remote 0x7fffea026580>, '__spec__': None, '__annotations__': {}, '__builtins__': <module at remote 0x7fffea15c290>, '__file__': '/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', '__cached__': None, 'np': <module at remote 0x7fffea0987d0>, 'LinearOperator': <type at remote 0x555555a8f710>, 'linear_operator_from_matrix': <function at remote 0x7fffea188940>, 'A': <numpy.ndarray at remote 0x7fff95382ec0>, 'B': <_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a52e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a5230>) at remote 0x7ff...(truncated), closeit=1, flags=0x7fffffffe268)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/pythonrun.c:1208
#23 0x00007ffff7d678da in _PyRun_SimpleFileObject (fp=0x555555588cf0,
filename='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', closeit=1,
flags=0x7fffffffe268) at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/pythonrun.c:456
#24 0x00007ffff7d66cee in _PyRun_AnyFileObject (fp=0x555555588cf0,
filename='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py', closeit=1,
flags=0x7fffffffe268) at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Python/pythonrun.c:90
#25 0x00007ffff7d98d97 in pymain_run_file_obj (
program_name='/home/andrey/Projects/scikit-learn/dev-env-debug/bin/python',
filename='/home/andrey/Projects/scikit-learn/linear-operator-matmul-fail.py',
skip_source_first_line=0) at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Modules/main.c:353
#26 0x00007ffff7d98e71 in pymain_run_file (config=0x55555555b680)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Modules/main.c:372
#27 0x00007ffff7d9958d in pymain_run_python (exitcode=0x7fffffffe3c4)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Modules/main.c:587
#28 0x00007ffff7d996c5 in Py_RunMain ()
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Modules/main.c:666
#29 0x00007ffff7d99783 in pymain_main (args=0x7fffffffe440)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Modules/main.c:696
#30 0x00007ffff7d9984b in Py_BytesMain (argc=2, argv=0x7fffffffe5a8)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Modules/main.c:720
#31 0x000055555555517d in main (argc=2, argv=0x7fffffffe5a8)
at /usr/src/debug/python3.10-3.10.7-1.fc36.x86_64/Programs/python.c:15PyArrayObject created (partial trace)#0 PyArray_NewFromDescr_int (subtype=0x7fffe9d277a0 <PyArray_Type>,
descr=0x7fffe9d2a260 <OBJECT_Descr>, nd=0, dims=0x7fffffffc580, strides=0x0, data=0x0,
flags=0, obj=0x0, base=0x0, zeroed=0, allow_emptystring=0)
at numpy/core/src/multiarray/ctors.c:693
#1 0x00007fffe9911cec in PyArray_NewFromDescrAndBase (subtype=0x7fffe9d277a0 <PyArray_Type>,
descr=0x7fffe9d2a260 <OBJECT_Descr>, nd=0, dims=0x7fffffffc580, strides=0x0, data=0x0,
flags=0, obj=0x0, base=0x0) at numpy/core/src/multiarray/ctors.c:958
#2 0x00007fffe9911c92 in PyArray_NewFromDescr (subtype=0x7fffe9d277a0 <PyArray_Type>,
descr=0x7fffe9d2a260 <OBJECT_Descr>, nd=0, dims=0x7fffffffc580, strides=0x0, data=0x0,
flags=0, obj=0x0) at numpy/core/src/multiarray/ctors.c:943
#3 0x00007fffe99139d9 in PyArray_FromAny (
op=<_CustomLinearOperator(dtype=<numpy.dtype[float64] at remote 0x7fffe9d29900>, shape=(3, 4), args=(), _CustomLinearOperator__matvec_impl=<function at remote 0x7fffe5ea6af0>, _CustomLinearOperator__rmatvec_impl=<function at remote 0x7fffe5d322b0>, _CustomLinearOperator__rmatmat_impl=<function at remote 0x7fff953a92e0>, _CustomLinearOperator__matmat_impl=<function at remote 0x7fff953a9230>) at remote 0x7fffea083b60>, newtype=0x0, min_depth=0, max_depth=0, flags=0, context=0x0)
at numpy/core/src/multiarray/ctors.c:1805
#4 0x00007fffe9ae0a92 in convert_ufunc_arguments (ufunc=0x7fffe6c58c00, full_args=...,
out_op=0x7fffffffc9b0, out_op_DTypes=0x7fffffffcab0, force_legacy_promotion=0x7fffffffc7cb "",
allow_legacy_promotion=0x7fffffffc7cc "\001", promoting_pyscalars=0x7fffffffc7cd "",
order_obj=0x0, out_order=0x7fffffffc7d0, casting_obj=0x0, out_casting=0x7fffffffc7d4,
subok_obj=0x0, out_subok=0x7fffffffc7ca "\001", where_obj=0x0, out_wheremask=0x7fffffffc7f8,
keepdims_obj=0x0, out_keepdims=0x7fffffffc7d8) at numpy/core/src/umath/ufunc_object.c:977
#5 0x00007fffe9aea9ed in ufunc_generic_fastcall (ufunc=0x7fffe6c58c00, args=0x7fffffffcef0,
len_args=2, kwnames=0x0, outer=0 '\000') at numpy/core/src/umath/ufunc_object.c:4896
#6 0x00007fffe9aeae8e in ufunc_generic_vectorcall (ufunc=<numpy.ufunc at remote 0x7fffe6c58c00>,
args=0x7fffffffcef0, len_args=2, kwnames=0x0) at numpy/core/src/umath/ufunc_object.c:5011 |
|
Does anyone have any general tips on debugging numerical issues? In my tests the outputs are "mostly" correct, but a few percent of the elements are off. I'm going to study the distribution of these errors a little bit and try to visualize what's happening. |
It's generally difficult. Debugging commonly requires math knowledge of numerical stability of the values examined and properties of algorithms involved. Start with everything in double precision and test in multiple environments with different algorithms to determine if inaccuracies are environment or algorithm dependent. |
Got it. What do you mean by an environment? |
|
Environment = different numerical compilers and libraries, e.g. provided by sklearn built-in PR testing. |
|
I see, thank you. Working on that now. |
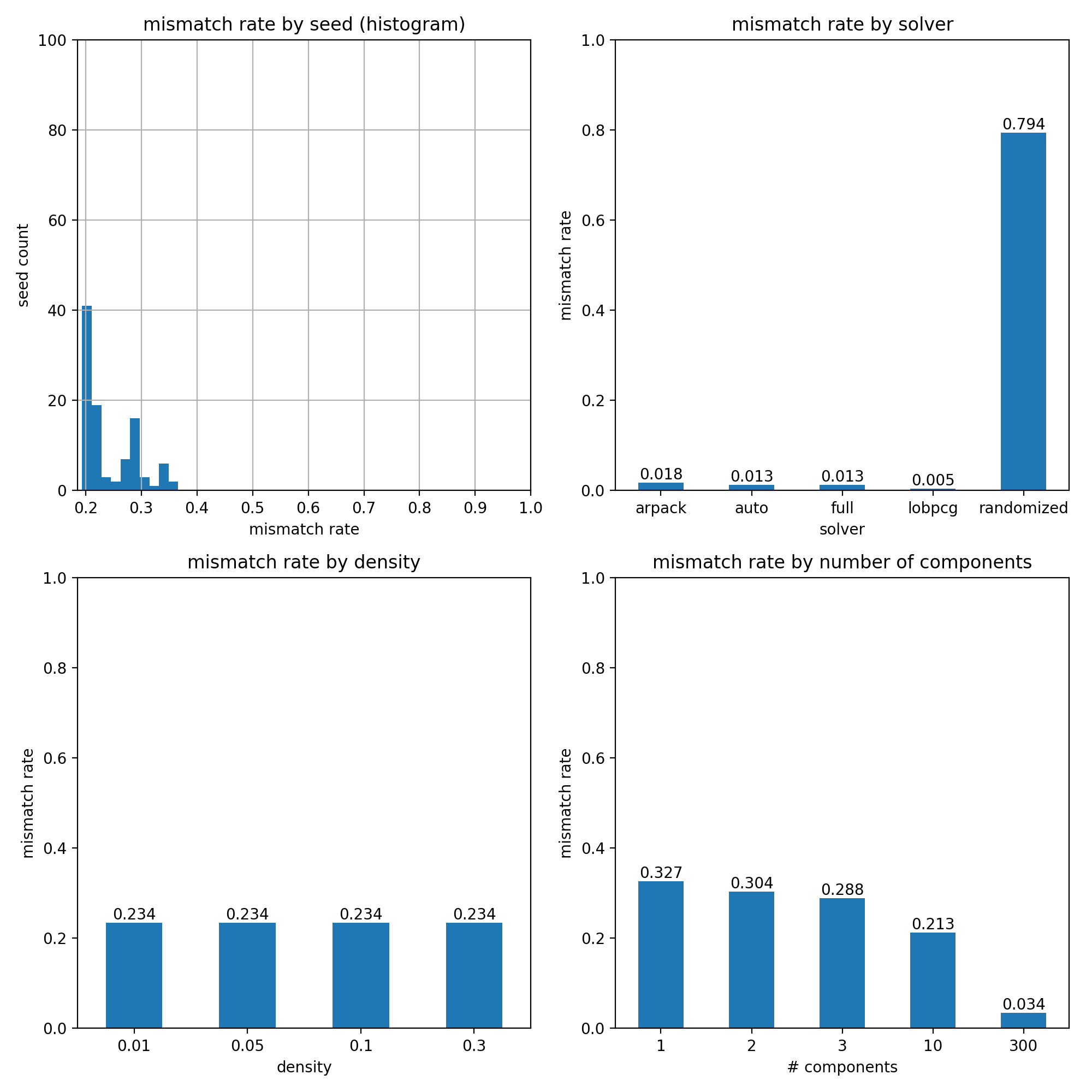
SummaryI added LOBPCG support and tested locally on all 100 global random seeds for a total of 18400 test cases. 14128 are failing, all failures are due to partial numerical mismatches. LOBPCG did pretty well, better than the default full solver. The randomized solver needs the most attention. Haven't added PROPACK yet. All tests were run using 64 bit floating point. Going to continue testing in other environments. Test matrixPCA_SOLVERS = ["full", "arpack", "randomized", "auto", "lobpcg"]
SPARSE_M, SPARSE_N = 400, 300 # arbitrary
SPARSE_MAX_COMPONENTS = min(SPARSE_M, SPARSE_N)
@pytest.mark.parametrize("density", [0.01, 0.05, 0.10, 0.30])
@pytest.mark.parametrize("n_components", [1, 2, 3, 10, SPARSE_MAX_COMPONENTS])
@pytest.mark.parametrize("format", ["csr", "csc"])
@pytest.mark.parametrize("svd_solver", PCA_SOLVERS)Pass rate by test parameterFraction of the tests that passed, broken down by test matrix parameter values. Elementwise mismatch rate by test parameterFor the tests that failed, the fraction of the elements that didn't pass the default |
|
Please have in mind that lobpcg internally calls the dense solver eigh if n_components > 5*min(N, M) |
Got it, thank you. I believe the test that I'm running shouldn't trigger the dense solver since I always have |
|
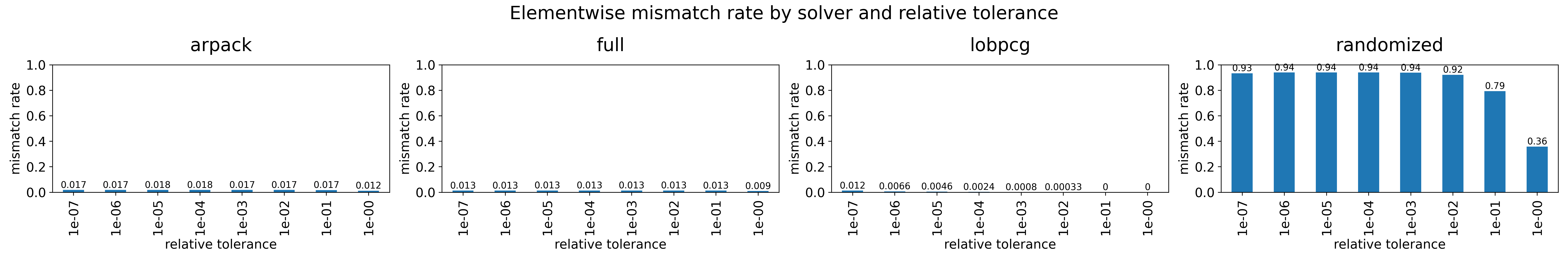
Given that most solvers have very few mismatched elements, I added I also triggered CI runs across the global random seed range (without the tolerance parameter) and now presumably have data from x86 and ARM Linux runs. Hope to visualize the data for both of the above next week. Will also need to compare intermediate outputs for the randomized solver since it has ~80% mismatches. Depending on how the tolerance study turns out, I will try to debug the numerical mismatches for the solvers other than the randomized, which definitely needs attention. |
Relative tolerance studyI varied the relative tolerance used in elementwise comparisons of the Below I'm showing plots of the resulting test pass rates and elementwise mismatch rates (of the failed tests) for each solver. Rate values are shown up to 2 significant digits. Tentative conclusionFor my personal practical purposes, LOBPCG passes the correctness test on this small data set (300x400). Don't know if those tolerances/error rates are acceptable in scikit-learn. ARPACK seems to always have a small number of elements that are significantly far away from the desired value, that could also be acceptable. Not sure yet how to debug. Randomized is performing very badly, even when tolerance is relaxed. I'll try to compare intermediate outputs to see where the divergence comes from. The full solver is not interesting for practical purposes. Pass rate by solverFraction of the tests that passed for each solver, broken down by relative tolerance level. Elementwise mismatch rate by solverFor the tests that failed, the fraction of the elements that didn't pass the comparison. |
|
I'm still trying to get a large number of tests to run across all environments on CI, I'll post an analysis of that later. It will likely only include the default tolerance level since the full range timed out. |
|
@andportnoy your tests results look plausible, except for the full solver which should be expected to be 100% accurate. |
|
@lobpcg Agreed, it's a little suspicious. I'll try to root cause. In each case I'm essentially comparing the SVD of the densified centered data to the SVD of the implicitly centered sparse data, using the same solver. Maybe I should instead use the outputs of a single, highest What do you think? |
|
You should probably use the full float64 solver as the baseline, after figuring our why it's not 100% accurate. |
|
Getting back to this. Short term goals:
|
|
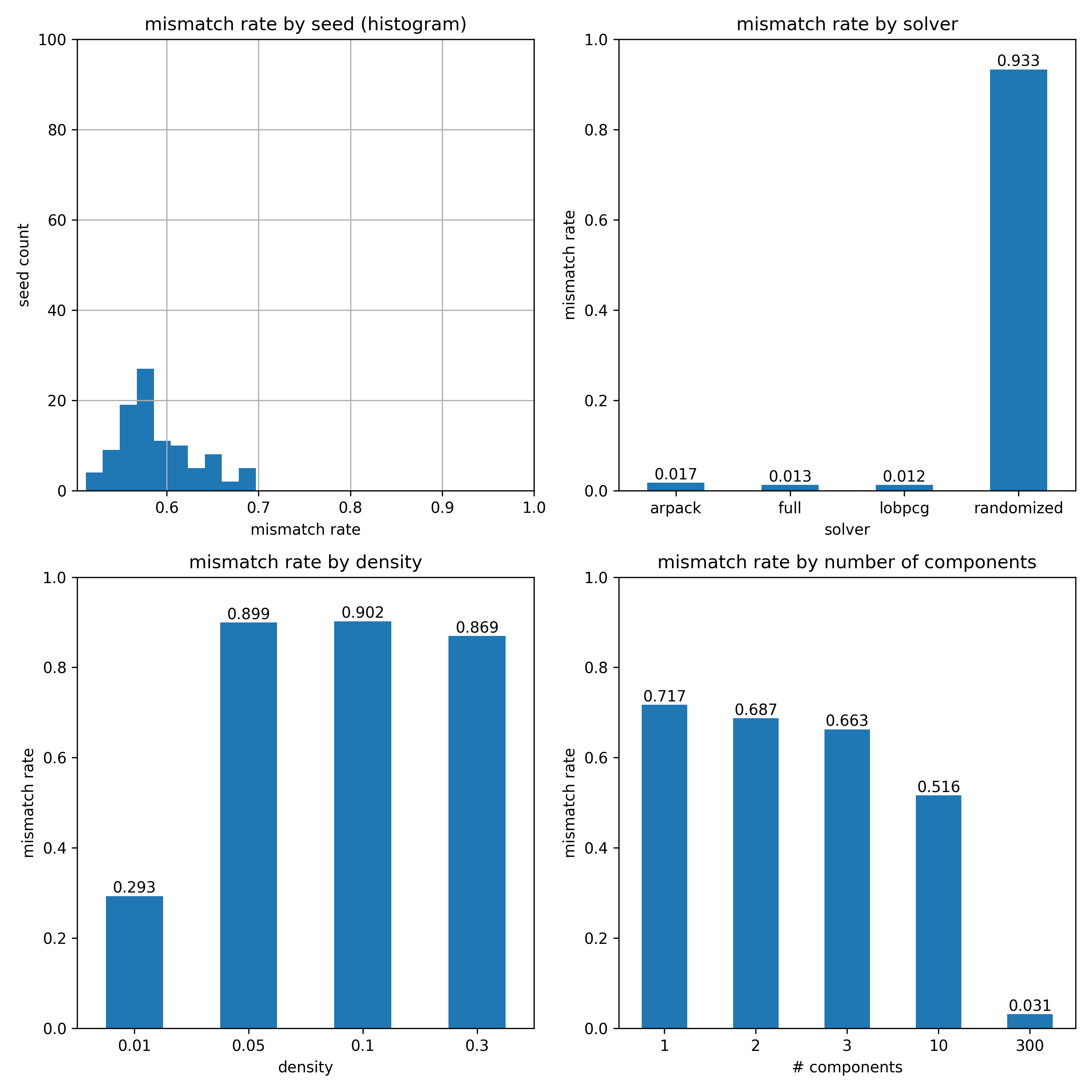
I discovered that I wasn't using the density parameter ( |
|
Plots for #12794 (comment) redone actually using the density parameter properly. Note the passrate is much better for higher densities. The plots can be reproduced by running bash pca-sparse-debug/scripts/passrate-main.sh
bash pca-sparse-debug/scripts/mismatch-main.shon the branch (commit 600f2f1). |
|
Plots for #12794 (comment) redone. |
|
Now need to get back to figuring out why the full solver has failures. |
Full solver debugSummaryDetailsAs is, 761/4000 (19.03%) full solver tests fail. These tests compare the following two outputs: A. densify the sparse matrix first, pass the result to the solver I've found some issues that are increasing the failure rate:
I don't know what role the distribution of our (randomly generated) inputs plays with respect to test outcomes. |
|
@andportnoy it appears that it might help your debugging if you run the extreme cases - examples with 1 or 2 rows or columns, where the svd part becomes trivial/exact so you would mostly check the operation of the mean. Your randomly generated input I guess is expected to always have an approximately constant (e.g., zero) mean, depending on a distribution. To change this, you may want to add random shifts to randomly generated columns/rows. |
|
One issue with the distribution here is that the entries are i.i.d., so the covariance matrix has a constant repeated on the diagonal and off diagonal entries all zeros… Would this be an "ill-conditioned" input to SVD, especially if the matrix is underdetermined? |





I suppose this is more of a feature request than anything else. There are several implementations of PCA that can compute the decomposition on noncentered, sparse data, while the implementation here does not support sparse matrices at all.
A matlab implementation can be found here and a Python implementation here. So far, I've been using the Python implementation, but it's missing some things and will eventually be deprecated (facebookarchive/fbpca#9).
I haven't looked at the code or math too much, but as far as I'm aware, it's just a matter of adding a term to
randomized_range_finderto account for centering.Is this something that you guys are aware of, and is anyone working on this? This would an awesome feature to have.
The text was updated successfully, but these errors were encountered: