New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
solve_ivp has a massive overhead #8257
Comments
|
Have you profiled the source to determine what it taking up the most time? |
|
iirc, this is mostly python per-call overhead, as per my last profiling.
The plan probably is to rewrite the innermost tight looping parts later.
1.1.2018 23.59 "xoviat" <notifications@github.com> kirjoitti:
… Have you profiled the source to determine what it taking up the most time?
—
You are receiving this because you are subscribed to this thread.
Reply to this email directly, view it on GitHub
<#8257 (comment)>, or mute
the thread
<https://github.com/notifications/unsubscribe-auth/AACI5iGyiur9zygE4tD7UPl6dMVBmhN2ks5tGVVOgaJpZM4RP7VC>
.
|
|
What can I say, it was written in Python relatively fast to provide features that people enjoy in Matlab. It is very far from ideal in efficiency for small problems, but I think it is sort of acceptable for typical scipy usage I think having code in Python also provides some value, it is easy to read and understand. Sure let's try to improve things step by step. For example, Runge-Kutta methods are quite straightforward and can be rewritten to compiled code relatively easy. |
|
I'm attempting to address this in gh-8386. However, I couldn't get the benchmark to work with either |
|
With SciPy 1.0 and other dependencies installed, I saw the following error: |
|
cc @Wrzlprmft Have you any idea why the benchmark isn't working? |
@xoviat: This is weird. Could be a problem of If this fails as well, can you try to run all tests of |
|
Result without lines 29-34: I'm running the tests now. |
|
@Wrzlprmft I'm struggling to create an environment that satisfies all of the dependencies required to run your script. I've attempted to build symengine.py from source to get ahead of the relevant issues, but have not been successful. Perhaps you could give some instructions to set-up the environment? |
|
@xoviat I never encountered problems with that or had any reports along that line (otherwise I would have left instructions in the documentation). Can you please open a separate issue here detailing your problems, so we do not have to digress too much here? My first guess is that the compilation fails but doesn’t throw an error for whatever reason. I vaguely recall that something like this happens with Jupyter and similar. Did you run it in a terminal? |
|
What I might do is wait for a symengine release and then come back to this. |
|
@Wrzlprmft hi, did you run the benchmark successfully? I run it with a problem of "TypeError: test_scenario() missing 6 required positional arguments: 'name', 'fun', 'initial', 'times', 'rtol', and 'atol'" Do you know why? Thank you |
|
@15940260868 This doesn’t happen to me and I really do not see why it should happen. It’s almost certainly not a problem of SciPy or JiTCODE. Did you make any changes to the code or run it in a weird way? |
|
Would be great to see For my problem (Lorenz system for 5000 time steps): Maximum difference in solutions: |
|
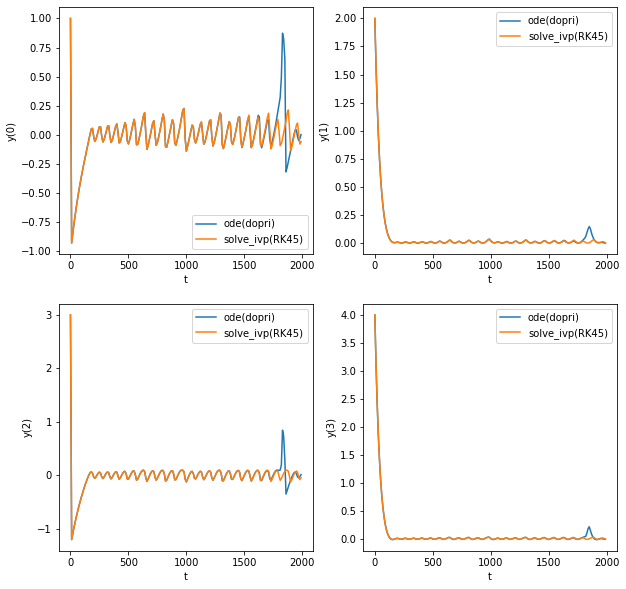
@billtubbs For LSODA, try using scipy-1.4.1 (should be fixed with gh-9901) I found that for the FitzHugh–Nagumo oscillators, dopri5 and solve_ivp(RK45) do not converge to the same solution. I tried looking at the trajectory (for time period 0-2000, instead of 2000-100000 as in the benchmark) and dopri5 shows a spike at around t=1800 before correcting itself to ivp(RK45)'s trajectory (but being late in phase) |
|
Thanks. I just upgraded to scipy-1.4.1 and here are the results (again, on my simulated Lorenz system for 5000 timesteps). Not much different: Scipy: 1.3.1
Scipy: 1.4.1
It's not a very accurate test (using B.t.w. I'm not concerned that they converge to different solutions. The Lorenz system is divergent. Code is here if you want to take a look. |
|
I wrote a wrapper to LSODA which has no overhead: https://github.com/Nicholaswogan/NumbaLSODA . Solving ODEs with this wrapper should be just as fast as pure C/C++. Below is a comparison with your sample lorenz system: Scipy: 1.5.2
"""Comparison of scipy's integration functions.
"""
import time
from functools import partial
import numpy as np
import scipy
from scipy.integrate import odeint, solve_ivp
from NumbaLSODA import lsoda_sig, lsoda
import numba as nb
def lorenz_odes(t, y, sigma, beta, rho):
"""The Lorenz system of ordinary differential equations.
Returns:
dydt (tuple): Derivative (w.r.t. time)
"""
y1, y2, y3 = y
return (sigma * (y2 - y1), y1 * (rho - y3) - y2, y1 * y2 - beta * y3)
@nb.cfunc(lsoda_sig)
def lorenz_odes_nb(t, y_, dy, p_):
y = nb.carray(y_,(3,))
p = nb.carray(p_,(3,))
y1, y2, y3 = y
sigma, beta, rho = p
dy[0] = sigma * (y2 - y1)
dy[1] = y1 * (rho - y3) - y2
dy[2] = y1 * y2 - beta * y3
funcptr = lorenz_odes_nb.address
print(f"Scipy: {scipy.__version__}")
dt = 0.01
T = 50
t = np.arange(dt, T + dt, dt)
# Lorenz system parameters
beta = 8 / 3
sigma = 10
rho = 28
n = 3
# Function to be integrated - with parameter values
fun = partial(lorenz_odes, sigma=sigma, beta=beta, rho=rho)
# Initial condition
y0 = (-8, 8, 27)
# Simulate using scipy.integrate.odeint method
# Produces same results as Matlab
rtol = 10e-12
atol = 10e-12 * np.ones_like(y0)
t0 = time.time()
y = odeint(fun, y0, t, tfirst=True, rtol=rtol, atol=atol)
print(f"odeint: {(time.time() - t0)*1000:.1f} ms")
assert y.shape == (5000, 3)
# Simulate using scipy.integrate.solve_ivp method
t_span = [t[0], t[-1]]
t0 = time.time()
sol = solve_ivp(fun, t_span, y0, t_eval=t, method='LSODA')
print(f"solve_ivp(LSODA): {(time.time() - t0)*1000:.1f} ms")
# Simulate using NumbaLSODA
data = np.array([sigma, beta, rho],dtype = np.float64)
y0_ = np.array(y0,dtype = np.float64)
t0 = time.time()
usol, success = lsoda(funcptr, y0_, t, data, rtol=rtol, atol=atol[0])
print(f"NumbaLSODA: {(time.time() - t0)*1000:.1f} ms") |
|
Apart from that, even though #8386 by 2018-@/xoviat was closed, I wonder if there's still interest in bringing it back to life? |
|
In an attempt to speed things up when working with Mentioning here because perhaps some of you might have hints how to approach the problem. In short, using Numba in unrelated code makes NaN value occur in the the BDF solver essentially at random. Never noticed so far on macOS; one documented case on Windows; easy to catch on Linux. The referenced Numba issue includes a minimal reproducer with a Dockerfile as well as a Github Actions CI set up for the very purpose of depicting the issue: https://github.com/slayoo/numba_issue_8931/actions/ Help needed :) |
|
@slayoo that's great to hear! |
While the new integrators or
solve_ivp, respectively, can compete withodefor large differential equations, it is up to twenty times slower for small ones, which suggests a massive overhead. This is not so nice, in particular considering thatodealready has a considerable overhead when compared toodeint(which mostly comes through the latter internalising the loops, as far as I can tell).Benchmark
The benchmark can be found in this Gist.
I here use my own module JiTCODE to compile the derivative to make things generally faster and emphasise what exactly is done by the benchmarked modules. Through this, the benchmark is not fair for
odeintsince the compiled derivative has to be wrapped to invert the arguments.On my machine this yields:
The text was updated successfully, but these errors were encountered: