New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ENH: override sf for rdist distribution #18586
Conversation
from scipy import stats
import numpy as np
from time import perf_counter
import matplotlib.pyplot as plt
rng = np.random.default_rng()
from mpmath import mp
mp.dps = 200
def rdist_sf_mpmath(x, c):
x = mp.mpf(x)
c = mp.mpf(c)
return float(mp.one - mp.betainc(c/2, c/2, 0, (x+1)/2, regularized=True))
def rdist_sf(x, c):
return stats.beta._sf((x+1)/2, c/2, c/2)
c = 541.0
x = np.logspace(-5, 10)
plt.loglog(x, stats.rdist.sf(x, c), label="rdist sf main", ls="dashed")
plt.loglog(x, rdist_sf(x, c), label="rdist sf pr", ls="dashdot")
plt.legend()
plt.show() |
|
It makes sense. Go ahead and revise the tests, and we can probably merge. (Would you also show the plot for arguments Alternatively, we could omit the custom test here. The generic tests confirms that this override is consistent with the rest of the distribution, and this is a very straightforward implementation of |
|
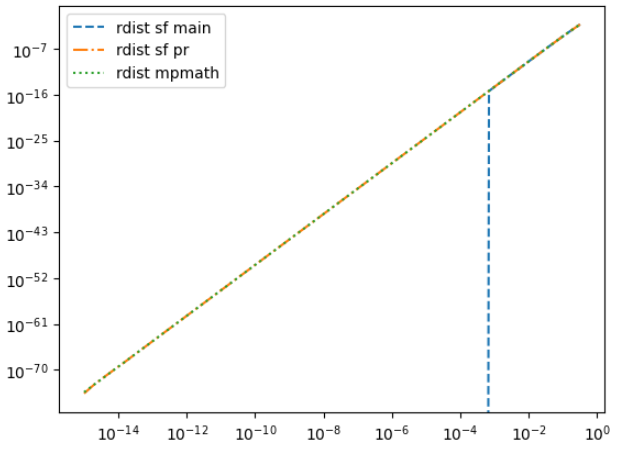
Here is the plot for c = 541.0
x = np.logspace(-15, -0.5)
x = 1 - x
mpmath_values = np.array([rdist_sf_mpmath(_x, c) for _x in x], np.float64)
plt.loglog(x, stats.rdist.sf(x, c), label="rdist sf main", ls="dashed")
plt.loglog(x, rdist_sf(x, c), label="rdist sf pr", ls="dashdot")
plt.loglog(x, mpmath_values, label="rdist mpmath", ls="dotted")
plt.legend()
plt.show() |
|
This is what i was going for. from scipy import stats
import numpy as np
from time import perf_counter
import matplotlib.pyplot as plt
rng = np.random.default_rng()
from mpmath import mp
mp.dps = 1000
def rdist_sf_mpmath(x, c):
x = mp.mpf(x)
c = mp.mpf(c)
return float(mp.one - mp.betainc(c/2, c/2, 0, (x+1)/2, regularized=True))
def rdist_sf(x, c):
return stats.beta._sf((x+1)/2, c/2, c/2)
c = 10
x = np.logspace(-15, -0.5)
mpmath_values = np.array([rdist_sf_mpmath(1-_x, c) for _x in x], dtype=np.float64)
plt.loglog(x, stats.rdist.sf(1-x, c), label="rdist sf main", ls="dashed")
plt.loglog(x, rdist_sf(1-x, c), label="rdist sf pr", ls="dashdot")
plt.loglog(x, mpmath_values, label="rdist mpmath", ls="dotted")
plt.legend()
plt.show()This shows that for moderate values of |
Nice! I understand, thank you for the explanation. Should I remove the tests that I added for this? |
|
I went ahead and checked the tests as they are. I think the first two would pass in main, so they aren't really needed, but having the second two can't hurt, even if they are mostly a test of the accuracy of float(mp.one - mp.betainc(c/2, c/2, (x+1)/2, mp.one, regularized=True))to float(mp.betainc(c/2, c/2, (x+1)/2, mp.one, regularized=True))because we might as well calculate the SF directly if we can. |



Reference issue
Towards gh-17832
What does this implement/fix?
Additional information
CC: @mdhaber Could you kindly have a look to see if this change makes sense? If it looks feasible then we can add the reference distribution to set up the tests.