Mabs is a genome assembly tool which optimizes parameters of genome assemblers Hifiasm and Flye.
The core idea of Mabs is to optimize parameters of a genome assembler to make an assembly where protein-coding genes are assembled more accurately than when the assembler is run with its default parameters.
Briefly, Mabs works as follows:
- It makes a series of genome assemblies by Hifiasm or Flye, using different values of parameters of these programs. Mabs uses several tricks to accelerate the assembly process.
- For each genome assembly, Mabs evaluates the quality of BUSCO genes' assembly using a special metric that I call "AG". For how AG is calculated, see calculate_AG.
- The genome assembly with the largest AG is considered the best.
Mabs is, on average, 3 times slower than Hifiasm or Flye, but usually produces better or equal assemblies. For details, see this article.
Mabs requires Python 3, Perl 5, GCC, Zlib-dev, Make.
Mabs should be installed in two steps:
- Install Python libraries Pandas, Plotnine, SciPy. This can be done, for example, by running the following two commands one after the other:
pip3 install --upgrade --user pip
pip3 install --upgrade --user Pandas Plotnine scipy - Download the latest version of Mabs from Releases. Extract the archive and run
bash install.sh
Alternatively, you can use a Singularity container with Mabs: https://mikeshelk.site/Diff/Mabs_distribution/Singularity_containers/ . To see information on how to use Mabs from the container, run a command "singularity run-help mabs.sif".
Two main components of Mabs are Mabs-hifiasm and Mabs-flye. Mabs-hifiasm works as a parameter optimizer of Hifiasm, while Mabs-flye works as a parameter optimizer of Flye.
Mabs-hifiasm is intended for PacBio HiFi (also known as CCS) reads. Also, it can be used for very accurate (accuracy ≥99%) Nanopore reads, as their characteristics are similar to characteristics of HiFi reads.
To run Mabs-hifiasm, a user should provide two values:
- A path to reads, via the option "--pacbio_hifi_reads".
- A BUSCO dataset. In the process of parameters optimization, Mabs uses a BUSCO dataset. The dataset can be provided using either the option "--download_busco_dataset", or the option "--local_busco_dataset". The option "--download_busco_dataset" requires a filename from https://mikeshelk.site/Data/BUSCO_datasets/Latest/ . It is recommended to use the most taxonomically narrow dataset. For example, if you assemble a drosophila genome, use "--download_busco_dataset diptera_odb10.2020-08-05.tar.gz". Alternatively, you can download a dataset to your computer manually, and use the option "--local_busco_dataset". For example, "--local_busco_dataset /home/username/Work/diptera_odb10.2020-08-05.tar.gz".
To see the full list of options, run
mabs-hifiasm.py --help
Since Mabs-hifiasm is based on Hifiasm (https://github.com/chhylp123/hifiasm), it can use paired-end Hi-C reads for haplotype phasing (but not for scaffolding). Provide trimmed Hi-C reads with options "--short_hi-c_reads_R1" and "--short_hi-c_reads_R2". If you want to do Hi-C scaffolding, you can use YaHS (https://github.com/c-zhou/yahs) with contigs made by Mabs.
Mabs-hifiasm can use ultra-long (N50 > 50 kbp) Nanopore reads to make assembly more contiguous. Provide them with the option "--ultralong_nanopore_reads".
Examples of using Mabs-hifiasm.
Example 1:
mabs-hifiasm.py --pacbio_hifi_reads hifi_reads.fastq --download_busco_dataset eudicots_odb10.2020-09-10.tar.gz --threads 40
Example 2:
mabs-hifiasm.py --pacbio_hifi_reads hifi_reads.fastq --short_hi-c_reads_R1 hi-c_reads_trimmed_R1.fastq --short_hi-c_reads_R2 hi-c_reads_trimmed_R2.fastq --ultralong_nanopore_reads ultralong_reads.fastq --download_busco_dataset diptera_odb10.2020-08-05.tar.gz --threads 40
Mabs-flye is intended for Nanopore reads and PacBio CLR reads (also known as "old PacBio reads"). Similarly to Mabs-hifiasm, Mabs-flye requires two values:
- A path to reads, provided via options "--nanopore_reads", "--pacbio_clr_reads" or "--pacbio_hifi_reads". If you have several read datasets created by different technologies, these options can be used simultaneously. Keep in mind that if you have only HiFi reads, it's usually better to use Mabs-hifiasm.
- A BUSCO dataset, provided via options "--download_busco_dataset" or "--local_busco_dataset". For details, see "2." in the description of Mabs-hifiasm above.
To see the full list of options, run
mabs-flye.py --help
Examples of using Mabs-flye.
Example 1:
mabs-flye.py --nanopore_reads nanopore_reads.fastq --download_busco_dataset eudicots_odb10.2020-09-10.tar.gz --threads 40
Example 2:
mabs-flye.py --nanopore_reads nanopore_reads.fastq --pacbio_hifi_reads pacbio_hifi_reads.fastq --download_busco_dataset diptera_odb10.2020-08-05.tar.gz --threads 40
Both Mabs-hifiasm and Mabs-flye have a similar output structure. Both of them create a folder which, by default, is named "Mabs_results". The name can be changed via the "--output_folder" option. The two main files that a user may need are:
- ./Mabs_results/mabs_log.txt
This file contains information on how Mabs-hifiasm or Mabs-flye run and whether they encountered any problems. - ./Mabs_results/The_best_assembly/assembly.fasta
These are the contigs you need.
If you are not sure whether Mabs-hifiasm and Mabs-flye have been installed properly, you can run
mabs-hifiasm.py --run_test
or
mabs-flye.py --run_test
These two commands assemble the first chromosome of Saccharomyces cerevisiae, which is approximately 200 kbp. If after the assembly finishes you see a file ./Mabs_results/The_best_assembly/assembly.fasta which is slightly larger than 200 KB, Mabs works correctly.
Besides Mabs-hifiasm and Mabs-flye, Mabs contains a third tool, named calculate_AG. Its purpose is to assess the genome assembly quality.
calculate_AG is used internally by Mabs-hifiasm and Mabs-flye, but also can be used externally if a user wants to assess the quality of some assembly.
The main concept in calculate_AG is "AG", which is the metric of gene assembly quality used by Mabs. "AG" is short for "the number of Accurately assembled Genes". It is calculated as the sum of the following two values:
a) The number of genes in single-copy BUSCO orthogroups.
b) The number of genes in true multicopy orthogroups. "True multicopy" means that there is more than one gene in these orthogroups not because of assembly errors, but because these genes are actual paralogs. In contrast, the number of genes in false multicopy orthogroups (the orthogroups where genes' duplications are because of assembly errors) is not included in AG.
AG, in my opinion, may be a better metric of gene assembly quality than BUSCO results, because BUSCO does not differentiate true multicopy orthogroups and false multicopy orthogroups, combining them into a single "D" category.
A frequent cause of false multicopy orthogroups are haplotypic duplications, when two alleles of a gene are erroneously assembled as paralogs.
calculate_AG differentiates true multicopy orthogroups from false multicopy orthogroups based on gene coverage, since if a duplication is an assembly error, the gene coverage should be decreased.
Basically, the larger AG is, the better the assembly is.
If a user wants to calculate AG for some genome assembly, he can use a command like:
calculate_AG.py --assembly contigs.fasta --nanopore_reads nanopore_reads.fastq --local_busco_dataset /mnt/lustre/username/Datasets/eudicots_odb10 --threads 40
For more options, run
calculate_AG.py --help
The main file produced by calculate_AG is ./AG_calculation_results/AG.txt . It contains a single number which is the AG.
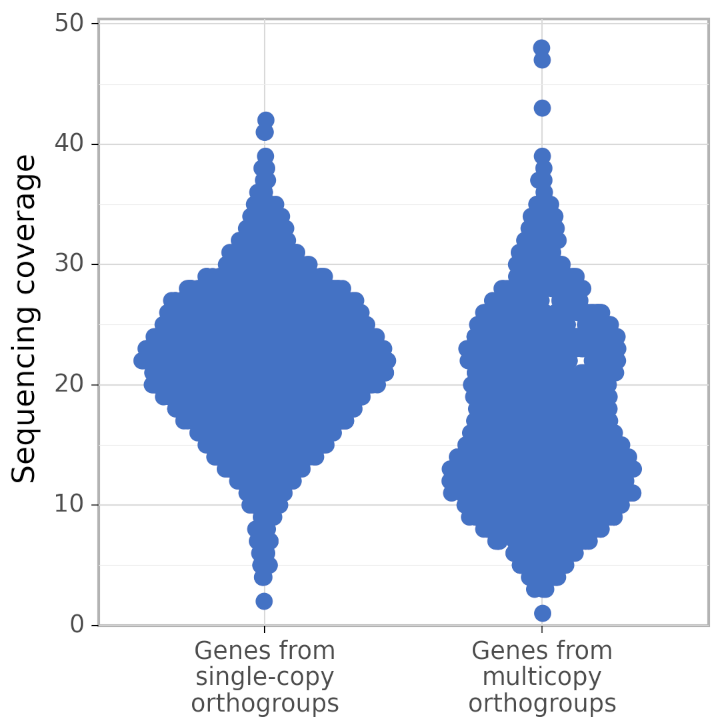
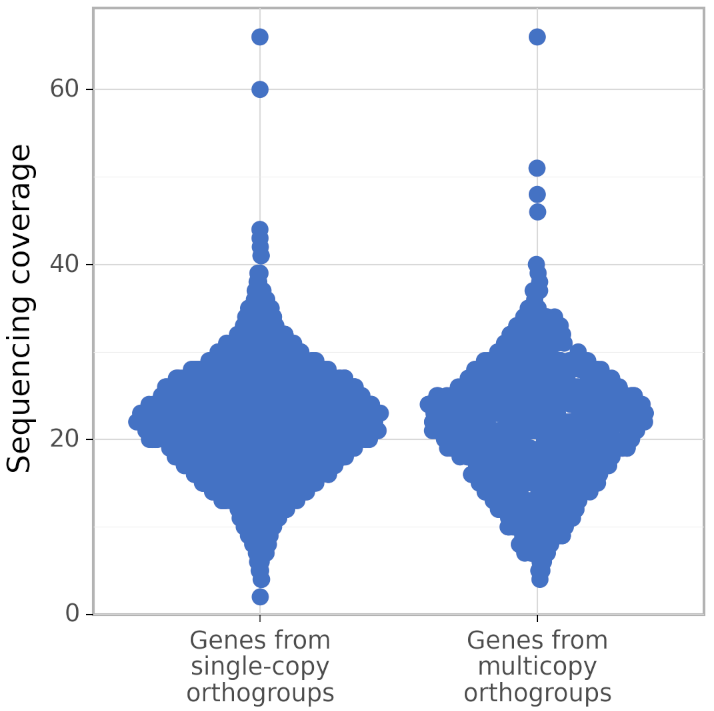
In addition, calculate_AG produces figures gene_coverage_distribution.svg and gene_coverage_distribution.png which look like this:
The right distribution usually has fewer genes than the left (because BUSCO orthogroups are usually single-copy). However, calculate_AG draws these two distribution such that they have the same area (but different density of points), to make a visual comparison of their shapes easier.
The recommended usage of calculate_AG is to compare the quality of assemblies of a single genome made by different genome assemblers, or made by a single assembler with different parameters. Besides the value of AG (in the file ./AG_calculation_results/AG.txt), calculate_AG also writes the exact numbers of genes in single-copy orthogroups, in true multicopy orthogroups, and in false multicopy orthogroups; the corresponding values can be found at the end of the file ./AG_calculation_results/log.txt.
- Should assemblies produced by Mabs be polished afterwards?
The assemblies made by Mabs-hifiasm are accurate already. The assemblies made by Mabs-flye require polishing by accurate reads. "Accurate reads" are reads of Illumina, MGI, or PacBio HiFi. Good programs for polishing are, for example, HyPo, POLCA, Racon. - How to assemble a genome using high-accuracy Nanopore reads?
If you have Nanopore reads with really high accuracy (≥99%), I advise trying both Mabs-hifiasm and Mabs-flye. - What is the program "Modified_hifiasm" used by Mabs?
Modified_hifiasm is a special version of Hifiasm, where I added an option "--only-primary". With this option, Modified_hifiasm stops after creating the file with the primary assembly. Usage of Modified_hifiasm makes Mabs-hifiasm faster than when using the original Hifiasm. - What is the main downside of Mabs?
I think, it is the fact that I made Mabs-flye instead of "Mabs-nextdenovo". When I started developing Mabs, I thought that Flye is the best assembler for Oxford Nanopore reads. However, I later realized that NextDenovo usually makes better assemblies. If I knew this beforehand, I would have made a version of Mabs based on NextDenovo instead of a version based on Flye. If you assemble a genome using Oxford Nanopore reads, I recommend to try both NextDenovo and Mabs-flye. - Is it worth using Mabs if I don't expect a high number of haplotypic duplications?
Though the primary purpose of Mabs is creation of assemblies with few haplotypic duplications, it may be useful even if you don't expect many haplotypic duplications. Since Mabs optimizes parameters of Hifiasm or Flye to maximize the gene assembly quality, in most cases it will produce assemblies better than or similar to Hifiasm or Flye. - Can Mabs be used to assemble metagenomes?
No. When evaluating which genes were assembled correctly and which were assembled incorrectly, Mabs relies on their coverage. In a metagenomic sequencing different genomes have different coverage, which makes Mabs useless. - Can Mabs be used to assemble haploid genomes, for example bacterial?
Yes. Though, I don't expect Mabs to be much better than Hifiasm and Flye for haploid genomes since haploid genome assemblies cannot have haplotypic duplications. - Can Mabs-hifiasm perform a trio binning assembly like Hifiasm?
Yes. You'll need to make "pat.yak" and "mat.yak" files as described in the readme of Hifiasm (https://github.com/chhylp123/hifiasm) and then provide them to Mabs via "--additional_hifiasm_parameters [-1 pat.yak -2 mat.yak]". - Should additional programs for haplotypic duplications removal (such as Purge_dups) be applied to assemblies made by Mabs?
In my experience, you can improve assemblies made by Mabs-flye by Purge_dups. However, in my experience, Purge_dups has detrimendal effect on assemblies made by Mabs-hifiasm. Still, you can try and see for yourself. - The option "--download_busco_dataset" fails to download a BUSCO dataset. What should I do?
This can happen if https://mikeshelk.site and, consequently, https://mikeshelk.site/Data/BUSCO_datasets/Latest/ is currently not accessible for some reason. To deal with this problem, manually download a file from http://busco-data.ezlab.org/v5/data/lineages/ and provide it to Mabs via the option "--local_busco_dataset". - Is the current algorithm of Mabs identical to the algorithm described in the article https://pubmed.ncbi.nlm.nih.gov/37794322/?
Not exactly. After the article was submitted to the journal, I made several changes in the algorithm. All of them are described in https://github.com/shelkmike/Mabs/releases . The most significant change was that the version of Mabs-flye described in the article assumed values of Flye parameters "assemble_ovlp_divergence" and "repeat_graph_ovlp_divergence" equal to each other; however, starting from the version 2.24, Mabs-flye optimizes values of these parameters independently (see https://github.com/shelkmike/Mabs/releases/tag/2.24) - What does "Mabs" mean?
Funny to say, but "Mabs" means "Miniasm-based Assembler which maximizes Busco Score". That's because:
a) The first version of Mabs was based on Miniasm instead of Hifiasm and Flye.
Miniasm takes as input a set of read overlaps produced by a program like Minimap2. Provided a file with overlaps, Miniasm performs assembly very quickly. The prominent speed of Miniasm allows exploring the parameter space more thoroughly than when using Hifiasm or Flye, which are 1-2 orders of magnitude slower. However, I later realized that the algorithm of Miniasm is inferior to the algorithms of Hifiasm and Flye, and even a more thorough exploration of a parameter space usually doesn't make Miniasm assemblies better than assemblies of Hifiasm and Flye. Therefore, I created a new version of Mabs that uses Hifiasm and Flye instead of Miniasm. The old Mabs optimized 4 parameters of Miniasm (in other words, "worked in a 4-dimensional parameter space"), while the current Mabs optimizes 1 parameter of Hifiasm and 2 parameters of Flye.
b) "Busco Score" is because very early versions of Mabs simply maximized BUSCO's "S" (the number of single-copy genes). However, I quickly realized that maximization of S may lead to collapsing of close paralogs, because it transfers them from the "multicopy" category to the "single-copy" category, thus increasing S. To deal with this problem, I started to classify multicopy genes into true multicopy (TM) and false multicopy (FM), and devised AG as a target for maximization, which is a sum of S and TM. - How to cite Mabs?
Cite the article https://pubmed.ncbi.nlm.nih.gov/37794322/.
In addition, if you used Mabs-hifiasm you may cite the article about Hifiasm (https://pubmed.ncbi.nlm.nih.gov/33526886/) and if you used Mabs-flye you may cite the article about Flye (https://pubmed.ncbi.nlm.nih.gov/30936562/) since Mabs is heavily based on these programs.