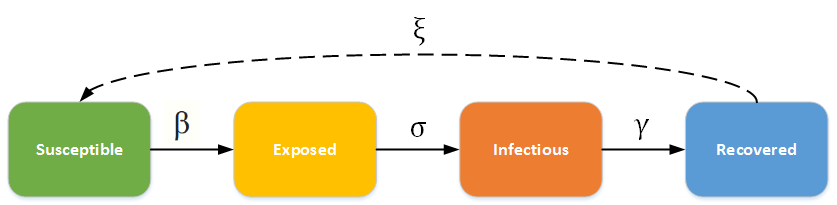
A minimal matlab class to generate SEIR epidemiology models that describe the
evolution of susceptible (S), exposed (E), infected (I), and (R) recovered populations.
This script can also fit the models to the infection data/agent based modeling
to obtain the model parameters and reproduction rate, R0.
Details on the models in this class along with the description of parameters can be found in IDM documentation
The following four models are included in the SEIRModel.m class:
closedSEIR: The classic SEIR model with four compartments and no births/deaths.vitalSEIR: SEIR Model that includes vital dynamics with equal the birth/death ratesclosedSEIRS: SEIR model in closed system with a fading immunity.vitalSEIRS: SEIR model with fading immunity and vital dynamics.
This model uses the matlab ODE function ode15s to solve for the time-dependent
evolution, and fsolve to compute the steady-state where applicable.
The data is fit using Parallel tempering algorithm or Simulated Annealing functions.
-
Equations: dynamic differential equations for the SEIR model. The differential equations describe the evolution of the infection with an incubation periodic. Four population compartments are chosen, namely, Susceptible (S), Exposed (E), Infected (I), Recovered (R). -
R0: Calculation of R0 for the given set of parameters. The reproduction number can be determined directly from the parameters. It can also be used to determine the herd immunity population. -
Equilibrium: Solves for the equilibrium condition. The SEIR models reach an endemic equilibrium at R0 > 1 and a disease free equilibrium at R0 < 1. One can find out by solving the equation for steady state. -
Dynamic: Solves for the time dependent condition. The SEIR model has a time dependent behavior where one can observe the evolution of Susceptible (S), Exposed (E), Infected (I), and Recovered (R) populations. -
FitData: Function that takes data and fits a SEIR(S) model. Obtain the model parameters and R0 for a time series data using a dynamic data fitting. The data needs to be in the format[time, S, E, I, R], where each entry is a column vector. -
plotDynamic: plots the time dependent evolution
| Parameters | Description |
|---|---|
N |
Total number of people |
mu |
Birth/death rate (assumed equal) |
beta |
Contact rate |
gamma |
Infection frequency (1/infection period) |
a |
Latent frequency (1/latent period) |
xi |
Lost immunity rate |
R0 |
Reproduction number |
modelType |
Type of model--closed/vital dyanmics SEIR(S) |