New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[Possible Bug] IntensityTable concatenate_intensity_tables() 'TAKE_MAX' strategy does not appear to be working quite right
#1432
Comments
|
I think your problem may be with the way you're setting up the physical coordinates on your IntensityTables using You should instead use |
|
thanks @njmei for this. I do expect the multi-overlap areas to be potentially inconsistent because the tile with the highest number of spots in the multi-overlap area might be different than the tile with highest number of spots in a pairwise overlapping area. This just comes with the territory of building the results out of pairwise take_max operations and it's a known limitation. I also think leaving the z coordinates out was a known limitation going into this, but the example above does look like a bug. (oops just saw @shanaxel42's comment.... I look forward to the resolution you two!) |
|
@shanaxel42 Thanks for the speedy reply! I'll try a EDIT: Are coordinates sorted by x then y? or y then x? |
ok so sorry @njmei I take back my previous comment about |
|
@shanaxel42 oh, well if it's helpful this version sorts by y_values then x_values but the problem still exists |
|
ok! Have done a bit more deep diving into this and I believe I understand what's happening. So one important thing to note is that when defining the boundaries of an IntensityTable during the overlap code I use the min/max values for x and y. So even though you may create an IntenistyTable with x coords in a range from 0 to 3, the min max values won't necessarily be 0 and 3. They are the min/max actual spot positions. For example the first IntensityTable you create |
|
Gotcha, many thanks for digging into this @shanaxel42! This is a bit different from how Brian had described this functionality to me... I replotted the example plot using the x,y min and max values to define boundaries as you described: It's clear now that the the blue square in the quad intersection, the two blue squares in the yellow/blue intersection, and the red star in the red/green intersection are literally on the boundaries, but are on outer boundary area for their 'zone' rather than the internal boundary. So those spots should probably be removed? The yellow star in the quad intersection is still a big ??? though. I'm going to defer to @berl on if this implementation is okay. |
|
@shanaxel42 On a related note, does the I see some discrepancies in the sum of Example:
|
|
the overlap code doesn't do anything special on a per round basis. It just compares two intensityTables based of physical coords. I'm guessing that the difference in spots has to do with the spot finding difference @ambrosejcarr mentioned this morning, the max_proj strategy? |
|
I guess what I'm wondering is: |
|
You understand it right, it selects/removes across the rounds. But maybe that's not the functionality you're looking for? The TAKE_MAX method was written specifically for you guys so if it's not working as expected that's completely my bad in misunderstanding your requirements. |
|
I think having any overlap strategy select/remove across rounds will result in overeager removal of spots. Not sure scientifically if this is a big deal, but this definitely would explain why I get discrepencies in number of CC: @berl |
|
Yeah, if it's done across all rounds, this strategy will result in some inconsistent results that we should avoid. This is probably on me- I should have been more careful when I first described this issue! The alignment within a round is assumed to be perfect here so all channels within a round can be treated together, but each round can have a different An important knock-on effect of this @njmei points out above: doing the |
|
Yes, it's definitely true that we're aiming for per-round processing. But we will need the ability to also do full experiment processing simply because there will be situations where already processed datasets will need to be revisited with tweaks to preprocessing/spotfinding hyperparameters (or maybe needing new algorithms applied). This is also why having different results when processing each individual round (as separate 'experiments') vs processing all rounds (as one 'experiment') is a big problem. |
|
@shanaxel42 would iterating over rounds and doing the |
When we figure out the difference, I think it would be a good idea for our tests to check for this. |
Upon reflection, am I correct that in the long term, we'd like to figure out how to resolve overlapping sections, which will require us to merge duplicated spots? If that's right, I think you have the right short term solution here, which is to add a flag that enables one to select the granularity over which the I don't have a great feeling for what the best default is. My best guess is that it's potentially per tile, if you want to err on the side of keeping data. |
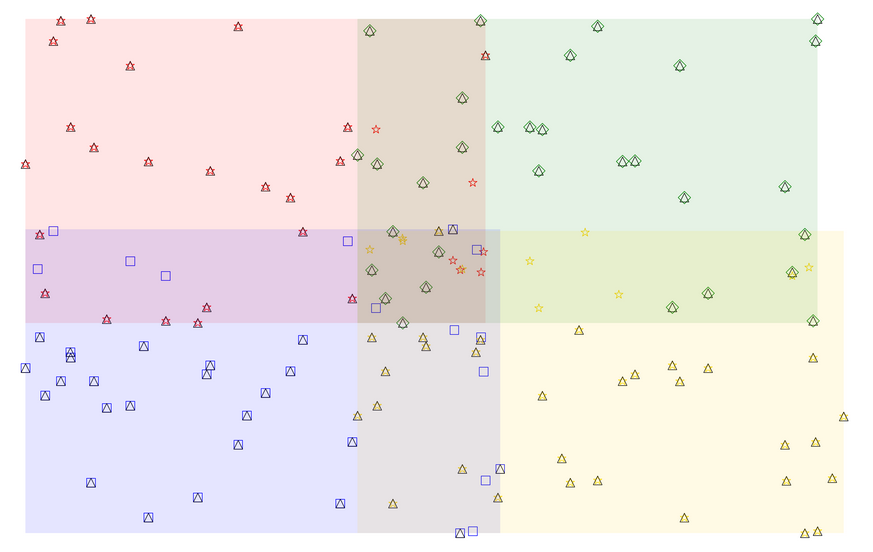
Some quick and dirty test code I wrote:
The weird result:
Black Triangles = spots in
concatenatedOther colored shapes = spots associated with a given color quadrant
The center quad overlap area shouldn't have that blue square or yellow star surviving in the concatenated table.
Also in the overlap area between yellow and blue, there are two anomalous blue spots that somehow survive the
TAKE_MAXprocess...Finally there is a surviving red star in the overlap between red and green.
Note also that this test code also doesn't even test for when Z-overlaps might occur.
CC: @berl
The text was updated successfully, but these errors were encountered: