-
Notifications
You must be signed in to change notification settings - Fork 214
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
KDTree for accelerating NMS #40
Conversation
…and passes them to nms implementation in stardist3d_impl; have not yet used the pairs in the nms implementation
…e - significantly faster
|
Oh I also just realized, the approximate times I give for the |
|
Hi Greg, thanks a lot for this contribution - it already looks very promising!
Thats a nice speed up! The 6Mio candidates are for the whole stack, I assume? So using I'm slightly worried about the additional memory footprint that a full kdtree of all candidates might bring - did you try measuring the peak memory usage? Additionally I suspect there is quite some heavy differences even for different kdtree implementations e.g. there are shows that i) there are quite some difference esp. for memory, and ii) the overall memory footprint is pretty high (e.g. 18GB for a mere 30k points for ckdtree). So having a similar plot as above but for memory consumption in your example would be nice and instructive. Additionally moving to |
|
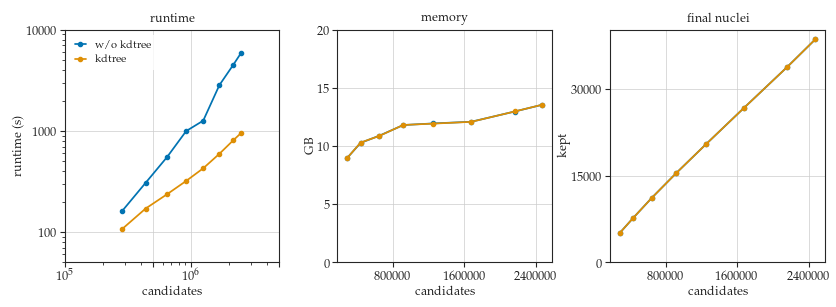
Hi Martin, Yes, the 6Mio candidates are for prediction on the whole stack. I did try For different kdtree implementations - I did look through this article written by the scikit-learn kdtree and balltree developer, but I didn't think there was enough differences in his graphs to justify introducing a new dependency (I don't think scikit-learn was a dependency before?) and I also preferred something with an underlying C++ implementation, as there are more big speed ups available if the kdtree data structure is available for query directly within the NMS algorithm. Unfortunately the only RAM measurements I have are the maximum and average usage (over the full run, prediction and NMS) for prediction on the full volumes both w/ and w/o the kdtree. The command in both cases was:
w/o kdtree max: 434.439GB w/o kdtree avg: 221.672GB I'm thinking that the kdtree and the query results for this many candidates took something like 70GB RAM. One thing that I haven't looked at in the code - it seems like the biggest expansion of data in the method is from raw data --> prediction; e.g. in my case I'm using 128 rays, so I need (129 * 2) more RAM relative to my input image (assuming 32 bit floats for probs and dists and 16 bit int for image). The n_tiles parameter cuts the image up into overlapping tiles for prediction. Does each tile also get separately prob thresholded before stitching back up for NMS? Based on the way RAM usage appears to me, it seems like no; but that would save a lot of space, since the dists of most voxels could be thrown out before the dists of new voxels were predicted/stored. I think if this were implemented, then the additional RAM of the kdtree would become an important factor. |
|
Hi Martin, I think you are right about scikit-learn kdtree outperforming scipy cKDTree. While working on another change to the package, I was led to trying the scikit-learn kdtree. For the same dataset considered above, the query was a few minutes faster than scipy cKDTree, but more importantly - due to the underlying data types used - the query results consumed significantly less RAM. For both packages I've found that the tree structure itself takes a negligible amount of RAM - I'm not sure exactly how this is implemented under the hood, but I suppose each node in the tree could be as small as a single pointer to where in the array the points are split. If that's the case you really only need something like 3*log_2(N) references (3 axes and N points). However it's implemented, empirically it's quite small. The query results however can be quite large, in the abstract it's a list of lists. The outer list is of length N (for N points) and the ith inner list is of length equal to the number of points within radius distance of the ith point. So this could be on the order of ~1000 * N long ints - that's something like 25GB without considering any extra metadata overhead. Scipy represents this object as a Python list of Python lists of Python ints. Python ints contain a cumbersome amount of metadata, so I don't think they're a great data type here. Scikit-learn on the other hand represents this as a Numpy array with So - combining the small query speed up and the better space efficiency, I've added code to use scikit-learn kdtree in a new commit. |
|
I've made some significant improvements by switching to the scikit-learn version of KDTree (good suggestion!) and changing the way the KDTree is used within NMS. To keep things clean, I'm going to delete my existing fork, create a new one, put the final version of the changes there, and submit them as (hopefully) a single commit. |
|
Hi @GFleishman, Thanks for all the work! Two things that I think are still worth pondering:
There likely won't be any difference between (Mean) and (Max) in practice (as your initial comparison showed), but I am still a bit wary of not having that guarantee.
So using (Max) will clearly take a hit on memory... |
|
Hi @maweigert, Right now I'm working on a different strategy from the software standpoint that will solve the extra time cost for using Max and the extra memory from having to store the larger lists of neighbors (potential suppression candidates). My first implementation in the PR was a good demonstration of kdtree, but definitely suboptimal w.r.t. what can be done with the Python C-API, which I'm learning on the fly so things have gone through various stages of kludge. The idea now is that I can call the sklearn kdtree object, and it's query_radius method, from within the C++ code itself - enabling a separate query for each polygon candidate just before the inner loop of NMS begins. That way, points which are "suppressed early" will never be queried against the tree - reducing total query time even if the distance threshold is large. The second benefit is that the code now only has to store one neighbor list at a time, thus reducing the total RAM used. This is definitely the best way to built it algorithmically but I still don't have the Python C-API down perfectly so implementing this today has been tricky. I also implemented what I mentioned before - applying the probability threshold for each tile separately during prediction. For the whole image, it cut max RAM down from ~400GB to ~50GB, and that was including the neighbors list from kdtree. So, if I can get the tile based threshold combined with the embedded python approach to the kdtree, I think I can get the whole zebrafish image segmented in like 10-15 minutes for something like 30GB RAM. Of course, the tile based prob threshold breaks a bunch of stuff, so I'll leave it to you guys to decide if you want to move in that direction generally or to just have my commit somewhere accessible on the repo so if others run into RAM issues there's some path to cutting it down without changing so many internal dependencies between functions. This is all WIP but I'm hoping to finish it soon. Thanks, |
|
Hi @GFleishman , sounds great!
Yes, that seems to be the preferable way. Having external source code be compiled and linked correctly via setup.py probably the biggest hurdle (it was quite some hassle with qhull).
Yes, I have a local branch that does exactly this. We haven't merged that feature yet, as long term we want to use the whole prob/stardist map for further shape refinement. But it might be worth reconsidering it for the pure stardist shapes (as it leads to huge memory reduction). Great to see this rolling! :) |
|
Hi @maweigert ,
Unfortunately I don't think I can do it this way, since sklearn kdtree is all scientific python stack under the hood, i.e. no C code. AFAIK you can't compile and link this like you would an external C library? My approach was to try and run sklearn kdtree from within the C code using the Python C-API - something like what's going on here. But it's been tricky to determine exactly what is needed, since this and the linked situation are different. Here we have a Python extension module (Stardist3d) which handles all the Python interfacing then calls implementations from another C++ file (Stardist3d_impl) passing only C style array pointers. In the linked code, they have an embedded Python interpreter which they interact with entirely within C. Although I see now it kind of mixes up the Python interface vs. implementation separation, I was trying to do things like: in Stardist3d_impl, which compiles with no warnings, but then the program just hangs on this line when executed - I guess it might eventually SegFault, but I never give it long enough to find out, after hanging for a few minutes I just kill the process, since that line shouldn't take longer than a few milliseconds(?). After trying multiple ways to initialize a numpy array in Stardist3d_impl and all showing the same failure mode, I'm trying to understand why this doesn't work, and also considering other designs like somehow having the kdtree object live in Stardist3d and just passing a callback function to stardist3d_impl or something. Do you have any ideas here?
Yeah, I figured that was the strongest pro for keeping the whole image of predictions. I think having a separate branch in the repo for RAM considerations is good though - then users can just switch and recompile to move into "maximum memory efficient" mode if that suits their application better. It could also eventually be part of the Config. So, I'll submit it on a separate branch/PR. This has been fun to work on so far :) |
|
Hi @GFleishman,
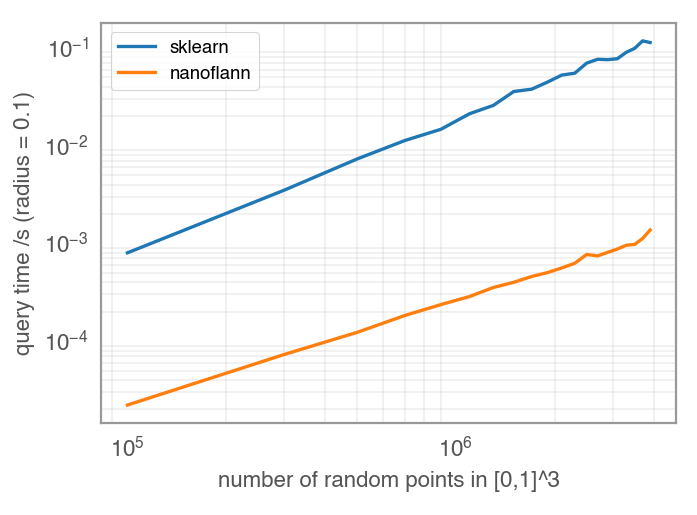
Yes, as far as I see incorporating sklearn into the c extension itself will become tricky and messy outright. There is another reason why this might not what we want: Currently, the c++ implementation ( But there might be good news! I just had a look into C++ kdtree implementations and there are some that look suitable for us. A good candidate might be nanoflann, which is BSD and even header only (meaning including a single hpp would be enough). There already is a radius query example which is exactly what we would need and it works just fine for me. Even better, some quick benchmark I did suggests, that query times are a magnitude faster than sklearn! So looks like a perfect match!
Yep, that is a sensible strategy.
It is! :) |
|
Hi @maweigert Awww - I just got the sklearn integration to work a few hours ago! There is a bit of trickyness involved in using the numpy C-API from multiple compilation units (here it would be stardist3d.cpp and stardist3d_impl.cpp); the answer there helped sort it out and got things working. I agree that keeping the Python module and C++ implementations separate is best. For now, the integrated sklearn strategy can at least demonstrate what kind of gains we'll get by putting the kdtree properly in the nms loop. I'm running some tests now and will report back. I did switch back to the "max" approach, but quite nicely I'm now using Great find with nanoflann! I'll have a look and see if I can get it to compile into stardist! Oh - did you see this note on the nanoflann readme about distances?:
Is it possible that you compared |
Makes sense!
Here's a short demo that shows how to perform a radius search (adapted from the nanoflann example): https://gist.github.com/maweigert/651355861950c5c310421e434ba3ceae Should be easy to adapt... |
Yes, that tripped me off at the beginning, but the other way around using radius^2=0.1 for nanoflann, so massively underestimating the speed up :) |
|
Great, thanks for the demo! In the meantime, I can confirm the expected speed up and RAM reduction for embedding the kdtree in the NMS loop. These are quick results from the same image I shared before: The NMS time was the same between embedded-kdtree and external kdtree, ~20 minutes However, the ~13 minutes required to query the tree for all candidate points is no longer present for the embedded-kdtree (since queries are done individually within the NMS outer loop). When combined with prob_thresholding on individual tiles, the maximum RAM over prediction and NMS for the embedded-kdtree was 17.378GB. |
|
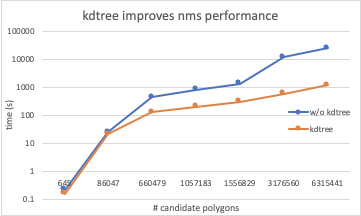
Hi, Following on this (great!) discussion , there are now two new branches
So while memory footprint is the same, the kdtree implementation is already 6x times for larger subvolumes. Pretty good! Note that this branch |
|
Hi again! |
|
Hello @maweigert! I hope the last 3 months have been good for you and that you're staying safe and sane. I've just gotten back into structural segmentation after a long trip doing functional segmentation. I've used the kdtree branch of the repo with nanoflann and it works very well. I haven't done any actual comparison, but the nms time seems as fast or faster than my implementation calling the sklearn methods using the python-API. One weird quirk I've noticed though - the NMS loop seems to run about 2x slower when I run So, I'm wondering where the prob/dist maps are after prediction on the gpu. Are these for sure sent back from gpu memory to system memory before the NMS loop starts? It seems like they for sure must be since they are C++ arrays/pointers but I don't know what I don't know so I thought I would ask before ruling this possibility out and looking further into potential differences on our cluster nodes. |
|
Hi @GFleishman
Good to hear! :)
This is strange and I don't see a reason why that would be the case.
Yes, they are in the CPU (they are returned by keras as numpy arrays). Before the NMS, both prob and dist are additionally converted to continuous c arrays, so they definitely are already on the host memory. How do you time the NMS loop? |
|
Hi @maweigert :),
This is what I suspected. Ok, I think it must be some difference between the gpu connected nodes and cpu exclusive nodes on our cluster. They are different processor models, so maybe I should try compiling on the gpu connected nodes? Previously I had compiled on one of the cpu exclusive nodes, so maybe it's more optimized for that processor architecture?
I'm running with |
Yes, that could be. I would suggest comparing CPU vs GPU speed on the very same GPU node and disable tf-gpu via |
|
@GFleishman FYI, the block-wise prediction in the latest release is now also shown here: https://github.com/mpicbg-csbd/stardist/blob/dev/examples/other2D/predict_big_data.ipynb |
This pull request solves issue #39.
After polygon candidates have been reformatted to parallel lists of 3D points and distances (sorted by probability), I construct a KDTree from the points using scipy.spatial.cKDTree. This takes negligible time for all of my tested cases (a few seconds at most). I then call kdtree.query_ball_tree which returns a list of lists - which is also parallel to points/dists. The i^th sublist contains the points which are sufficiently close to point i to merit comparison in NMS. This query is somewhat time consuming, for the largest case tested (6.32e6 points) it took 23 minutes.
This list of lists is passed to
c_non_max_suppression_indsusing theO&option toPyArg_ParseTupleand a custom python -> c data types converter functionlists_to_vectors.The inner loop of
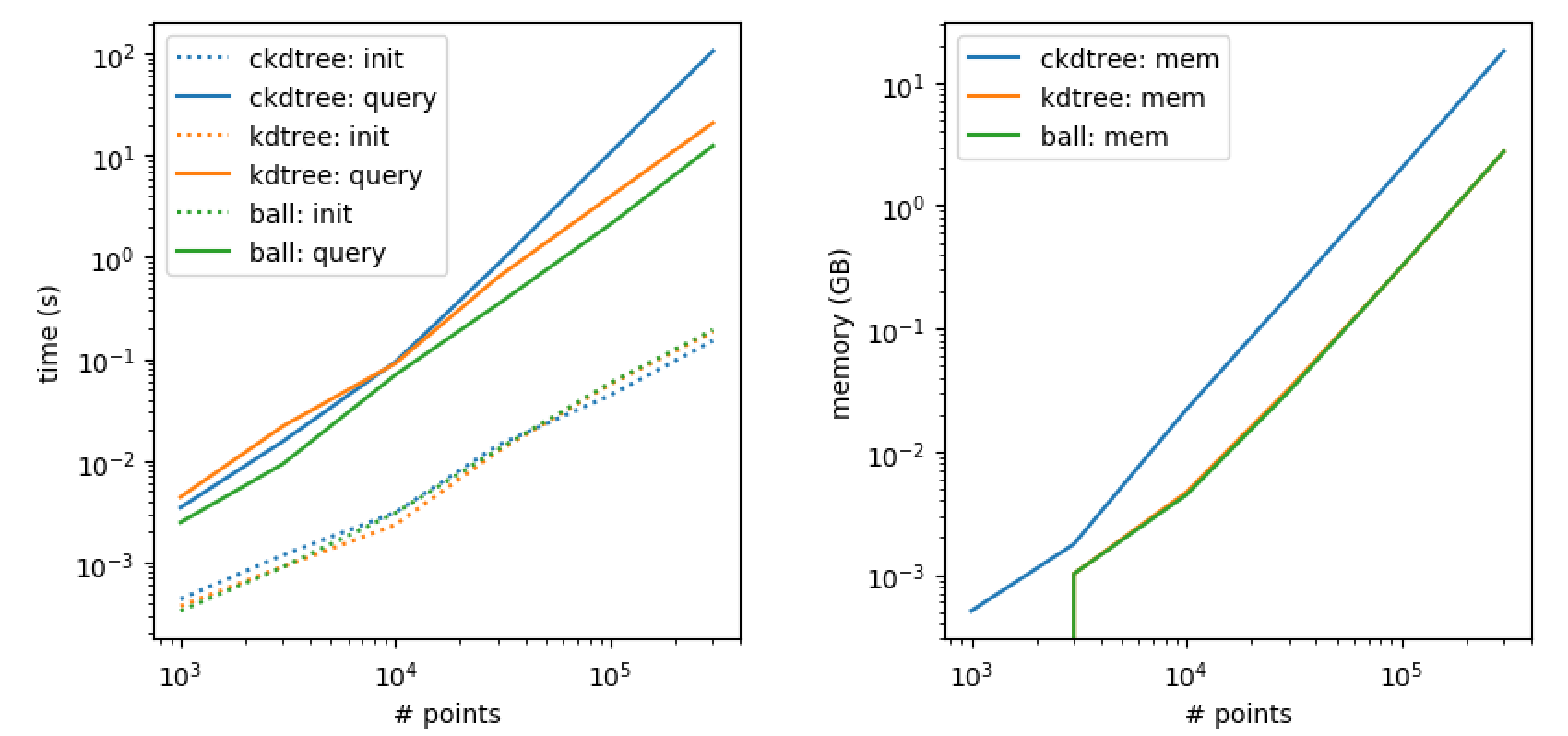
_COMMON_non_maximum_suppression_sparseis modified to only iterate over these candidate pairs. This capitalizes on the investment of the previous bounded neighbors query. For the largest tested case it brought NMS time down from 7.5 hours to 20 minutes, while still finding the same set of polygons.I evaluated NMS time over a wide number of polygon candidates. Here are the results (note the log_10 scale on the vertical axis):
In all cases except the largest, the number of detected polygons between the baseline method and using the kdtree was exactly the same. For the largest case, the baseline method detected 90535 polygons and the kdtree method detected 90536 polygons. Here is a side by side comparison of a single plane of polygons detected in the largest case for both methods:
and an overlay (baseline method in red, with kdtree in green):
Correspondence is exact through the whole stack - I'm not sure where that extra one is - too hard to find by eye :) !
Some additional notes
The kdtree query time also scales with number of points, but with a worst case complexity of O(N*logN). For very few points (left side of plot) the query time is negligible (milliseconds) - so the kdtree doesn't hurt in those cases.
The O(NlogN) query time for the kdtree could also be reduced to O(ClogN) where C is a constant strictly smaller than N, using a better implementation. Currently, I'm building the kdtree in python, running the bounded neighbors query for all points and then passing the result to the C++ code. Some of these points get suppressed before their bounded neighbors lists are used, and thus computing their bounded neighbors was a waste. If the kdtree data structure was directly accessible in the C++ code, you would instead want to query for bounded neighbors of each individual point at the start of the inner loop of NMS. That way, points which were already suppressed never need to be queried; I think this would cut the query time in half at least. It's a bit more tricky to implement, but definitely possible. I would probably grab the cKDTree implementation from scipy and start from there.
The one extra polygon detected for the largest test case illustrates an interesting aspect of the kdtree.query_ball_tree (bounded neighbors) search. You must provide an upper bound. You can guarantee that any polygon which could be suppressed by a point is included in it's bounded neighbors list by using
2 * np.max(dists)as the upper bound (any polygons further than that can not possibly overlap), but this results in a pretty long query time, though likely still an overall gain compared to NMS w/o the kdtree. The current choice is2 * (np.mean(dists) + np.std(dists))which gives acceptable results for my dataset and is a bit quicker than the max - so I've stuck with it.