Repository containing the work done for my Großer Beleg on "3D Medical Image Fusion using Convolutional Neural Networks" at Technische Universität (TU) Dresden. Fork of TFGAN with a fix for mode collapse by skipping the opacity GAN. This also enables generalisation of the network to work with any arbitrary 3D volume instead of only the one it was trained with. Also updated for PyTorch 1.3.1 (was 0.2) with support for the NIFTI file format (Volumetric Scalar Arrays).
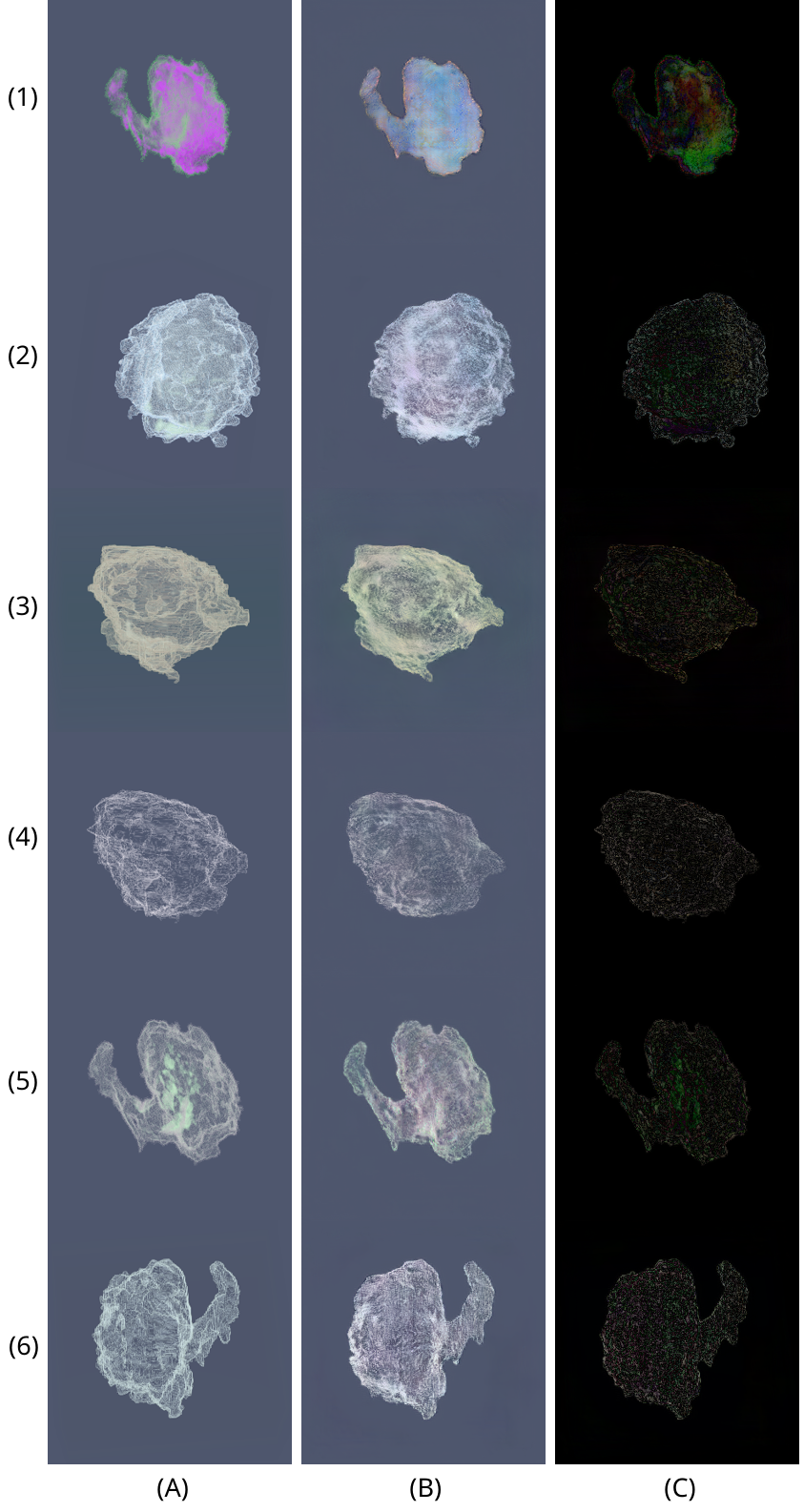
Qualitative comparison of 6 volume renderings of the volume the network was trained on. Groundtruth (A), prediction (B) and difference (C).
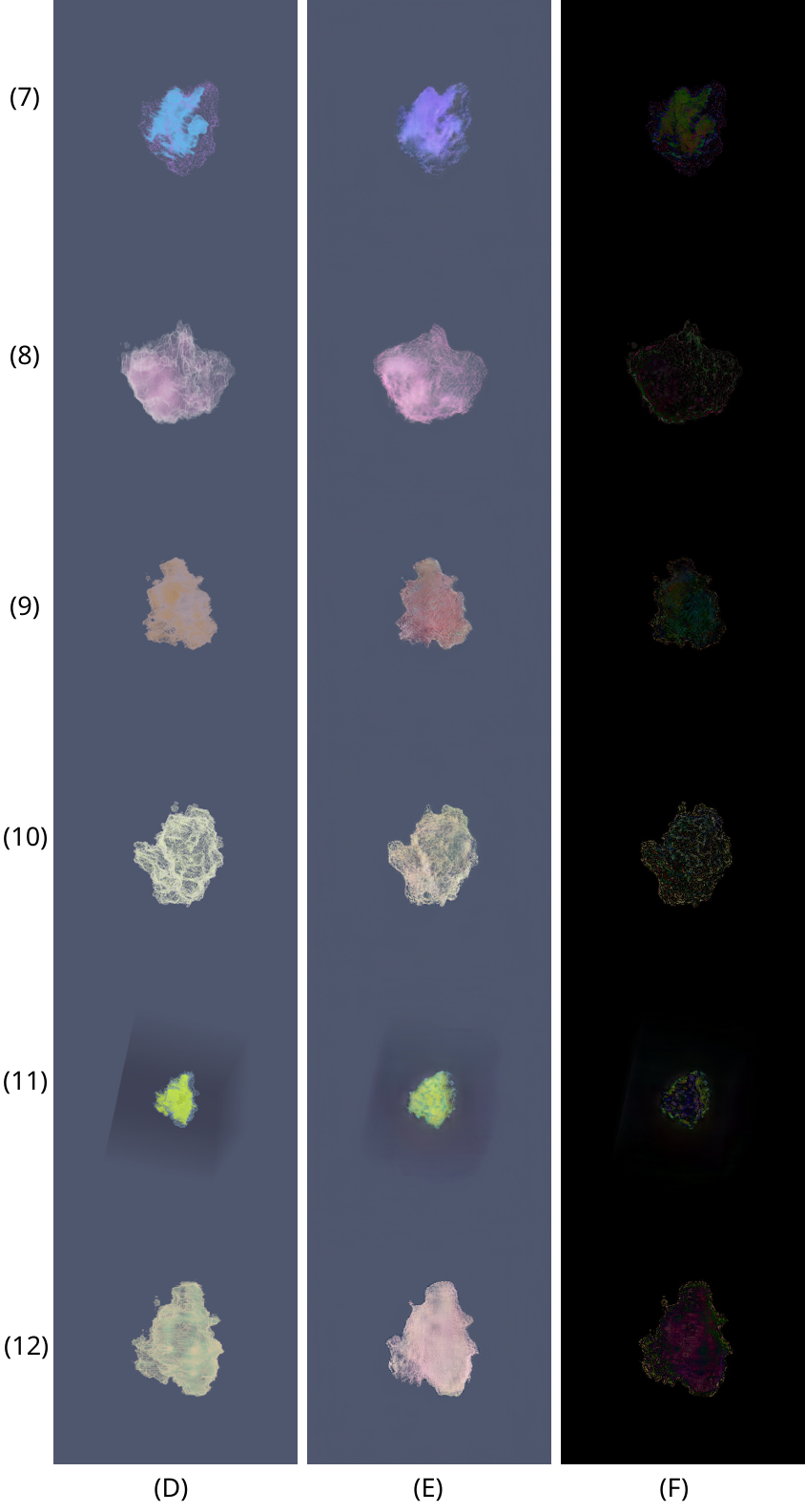
Qualitative comparison of 6 volume renderings of a new volume unknown to the network. Groundtruth (D), prediction (E) and difference (F).
We have successfully shown that our method is quite robust in replacing the traditional volume rendering pipeline and could be used as a real-time application to generate quality volume rendered tumour images. One of the main contributions is the robust replacement of a complex volume rendering pipeline through a simpler, faster GAN-based pipeline. Our trained GAN network can generate volume rendering predictions of any 3D dataset, given the viewpoints, opacity and colour transfer functions. Our proposed results exceed those of TFGAN, which suffered from mode collapse in the opacity GAN stage. Quantitative and qualitative performance measured with RMSE, EMD and MS-SSIM was generally good, except for some artifacts and errors encountered on a few outliers. However, most importantly and contrary to the original TFGAN paper, our network is able to generate volume renderings not just of the volume it was trained on, but of any reasonably similar volume. The generalisation of a GAN-based volume rendering method makes it a lot more useful and applicable to a wider category of 3D volumes outside of medical tasks.
While we trained the network on just one volume, it is probable that overall performance would increase if it was trained on a larger number of samples generated from multiple volumes. We estimate that transfer function performance would increase, although this might be at the cost of a diminished regard for depth and details. Unfortunately, verifying this hypothesis and trying to improve our results further would have been difficult because of the time and technical constraints caused by the usage of a personal computer for the network’s training.
In the future, our approach could be extended to add interactivity to the network through a user interface similar to the one used in TFGAN, to give the user access to categories of new transfer functions for varying types of volume visualisations.
This software was tested using the following packages and versions:
| Software | Version |
|---|---|
| Python | 3.7.3 |
| PyTorch | 1.3.1 |
| numpy | 1.16.4 |
| scikit-image | 0.16.2 |
| Pillow | 5.3.0 |
| nibabel | 2.5.1 |
| jupyterlab | 1.2.2 |
-
Generate n samples of random metadata (viewpoints, opacity and color transfer functions):
python render_random_meta.py volume.nii output_path n -
From the metadata, generate volume renderings:
python render_volume_images.py volume.nii output_path/view.npy output_path/opacity.npy output_path/color.npy -
Train the network for x epochs:
python stage2_gan.py --dataroot output_path --outf checkpoint_path --checkpoint_dir checkpoint_path --niter x